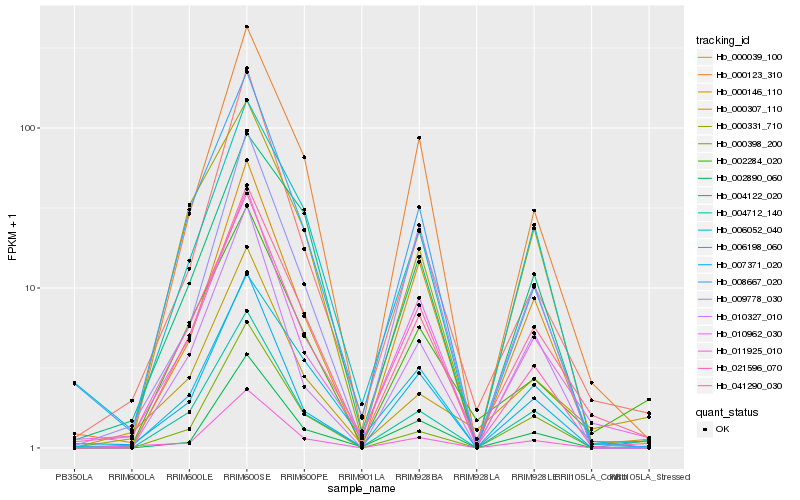
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_041290_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 2 |
Hb_004712_140 |
0.087365973 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 3 |
Hb_000123_310 |
0.1035657998 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 4 |
Hb_010962_030 |
0.1045598162 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_006198_060 |
0.1086824941 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 6 |
Hb_006052_040 |
0.1179396451 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 7 |
Hb_000146_110 |
0.120343504 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 8 |
Hb_021596_070 |
0.1210953943 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_000331_710 |
0.1313784075 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_002284_020 |
0.1332934016 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 11 |
Hb_000398_200 |
0.1336598715 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 12 |
Hb_002890_060 |
0.1355923395 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 13 |
Hb_000039_100 |
0.1369349711 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 14 |
Hb_007371_020 |
0.1387855225 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_011925_010 |
0.139830991 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008667_020 |
0.1418936406 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_010327_010 |
0.1428340519 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_000307_110 |
0.143853636 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 19 |
Hb_004122_020 |
0.1441865428 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 20 |
Hb_009778_030 |
0.1447428033 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |