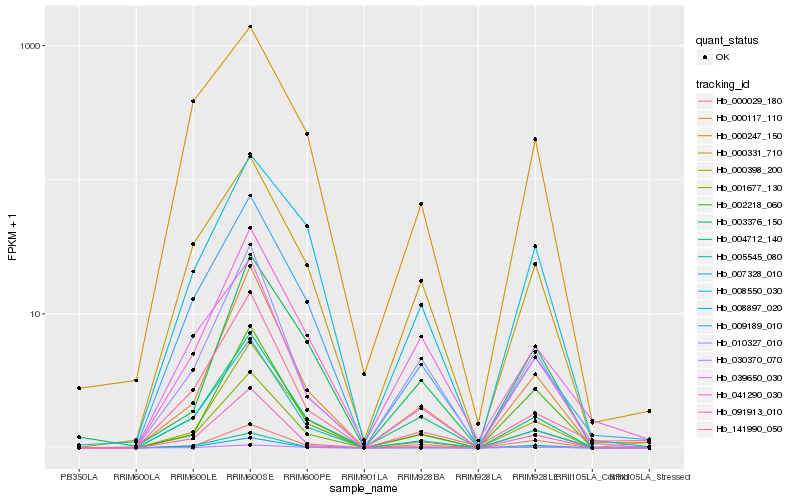
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001677_130 |
0.0 |
- |
- |
hypothetical protein POPTR_0081s002101g, partial [Populus trichocarpa] |
| 2 |
Hb_000398_200 |
0.0537477088 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 3 |
Hb_000117_110 |
0.0752129866 |
- |
- |
transferase, putative [Ricinus communis] |
| 4 |
Hb_007328_010 |
0.1021464035 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 5 |
Hb_004712_140 |
0.1085355451 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 6 |
Hb_091913_010 |
0.1127343823 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 7 |
Hb_010327_010 |
0.1233049308 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_002218_060 |
0.129017684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_030370_070 |
0.1314765248 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_141990_050 |
0.1389696001 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 11 |
Hb_000331_710 |
0.1389881447 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_009189_010 |
0.139390337 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 13 |
Hb_039650_030 |
0.1409658721 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 14 |
Hb_005545_080 |
0.1435262091 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 15 |
Hb_008897_020 |
0.1465885336 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Musa acuminata subsp. malaccensis] |
| 16 |
Hb_003376_150 |
0.1478897035 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 17 |
Hb_041290_030 |
0.1479159182 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 18 |
Hb_008550_030 |
0.1488815745 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |
| 19 |
Hb_000029_180 |
0.1506670868 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 20 |
Hb_000247_150 |
0.1537364812 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |