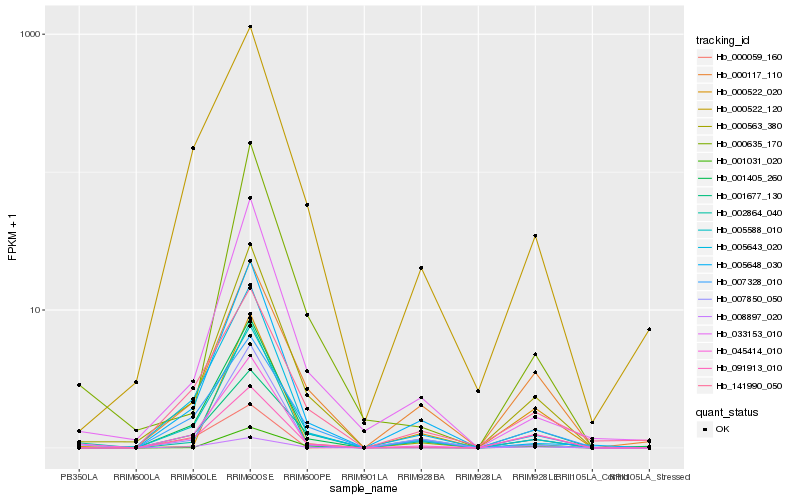
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_045414_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000563_380 |
0.1215709228 |
- |
- |
hypothetical protein CICLE_v10007206mg [Citrus clementina] |
| 3 |
Hb_091913_010 |
0.1332282146 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 4 |
Hb_008897_020 |
0.1434064593 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_005643_020 |
0.1454375808 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 6 |
Hb_005588_010 |
0.1464736987 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 7 |
Hb_002864_040 |
0.1494672788 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 8 |
Hb_007850_050 |
0.1571637473 |
- |
- |
flavonoid 3-hydroxylase, putative [Ricinus communis] |
| 9 |
Hb_007328_010 |
0.1580298126 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000522_120 |
0.1659937712 |
- |
- |
hypothetical protein POPTR_0008s16470g [Populus trichocarpa] |
| 11 |
Hb_001405_260 |
0.1682130707 |
- |
- |
E3 ubiquitin ligase PUB14, putative [Ricinus communis] |
| 12 |
Hb_033153_010 |
0.1774785349 |
- |
- |
PREDICTED: MLO-like protein 12 [Jatropha curcas] |
| 13 |
Hb_141990_050 |
0.1803232894 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 14 |
Hb_000117_110 |
0.1835761225 |
- |
- |
transferase, putative [Ricinus communis] |
| 15 |
Hb_000522_020 |
0.1876525768 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 16 |
Hb_001677_130 |
0.1894893162 |
- |
- |
hypothetical protein POPTR_0081s002101g, partial [Populus trichocarpa] |
| 17 |
Hb_005648_030 |
0.1910696541 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH18-like [Jatropha curcas] |
| 18 |
Hb_000059_160 |
0.1911672339 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 19 |
Hb_001031_020 |
0.1915263468 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 20 |
Hb_000635_170 |
0.1940898608 |
- |
- |
cytochrome P450, putative [Ricinus communis] |