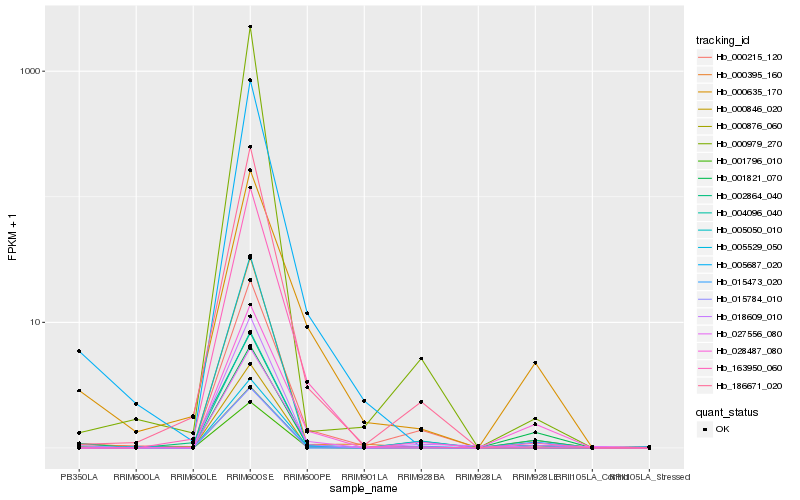
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000876_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000846_020 |
0.07450703 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_004096_040 |
0.088872054 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_002864_040 |
0.0985352431 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 5 |
Hb_005529_050 |
0.0997538406 |
- |
- |
PREDICTED: cytochrome P450 94B3-like [Nelumbo nucifera] |
| 6 |
Hb_015473_020 |
0.1152985508 |
- |
- |
hypothetical protein B456_001G090400 [Gossypium raimondii] |
| 7 |
Hb_001821_070 |
0.1180490914 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |
| 8 |
Hb_028487_080 |
0.1203091131 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 9 |
Hb_015784_010 |
0.1260690639 |
- |
- |
hypothetical protein VITISV_019364 [Vitis vinifera] |
| 10 |
Hb_000395_160 |
0.1269205506 |
- |
- |
hypothetical protein POPTR_0006s25490g [Populus trichocarpa] |
| 11 |
Hb_005687_020 |
0.1269460517 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 12 |
Hb_000979_270 |
0.1301444417 |
- |
- |
PREDICTED: MLP-like protein 31 [Jatropha curcas] |
| 13 |
Hb_186671_020 |
0.1316832384 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_001796_010 |
0.1323100021 |
- |
- |
purine transporter, putative [Ricinus communis] |
| 15 |
Hb_027556_080 |
0.135269988 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g24130 [Jatropha curcas] |
| 16 |
Hb_005050_010 |
0.1368068103 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 17 |
Hb_018609_010 |
0.1396808182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000215_120 |
0.1412863253 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 19 |
Hb_000635_170 |
0.1415198354 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_163950_060 |
0.1415438071 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |