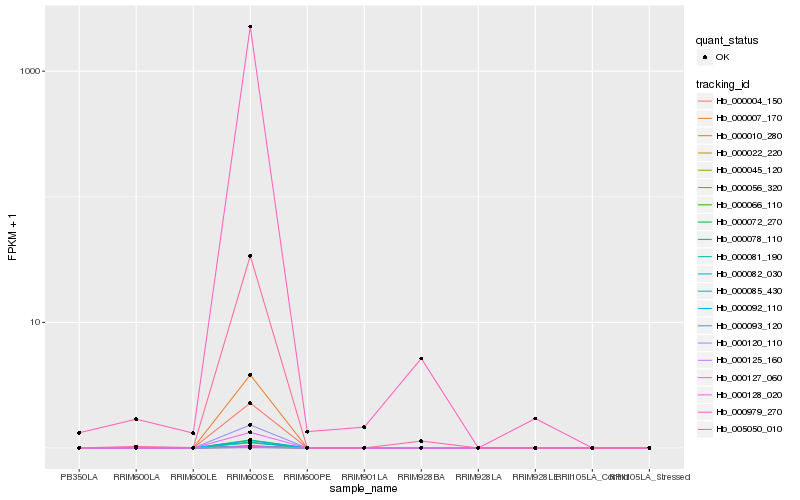
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000979_270 |
0.0 |
- |
- |
PREDICTED: MLP-like protein 31 [Jatropha curcas] |
| 2 |
Hb_005050_010 |
0.0287315442 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 3 |
Hb_000004_150 |
0.0392920744 |
- |
- |
hypothetical protein RCOM_1382630 [Ricinus communis] |
| 4 |
Hb_000007_170 |
0.0392920744 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 2-like protein [Nelumbo nucifera] |
| 5 |
Hb_000010_280 |
0.0392920744 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 6 |
Hb_000022_220 |
0.0392920744 |
- |
- |
PREDICTED: potassium channel KAT3 [Jatropha curcas] |
| 7 |
Hb_000045_120 |
0.0392920744 |
- |
- |
hypothetical protein JCGZ_01307 [Jatropha curcas] |
| 8 |
Hb_000056_320 |
0.0392920744 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000066_110 |
0.0392920744 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01370 [Jatropha curcas] |
| 10 |
Hb_000072_270 |
0.0392920744 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 11 |
Hb_000078_110 |
0.0392920744 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 12 |
Hb_000081_190 |
0.0392920744 |
- |
- |
- |
| 13 |
Hb_000082_030 |
0.0392920744 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105164525 [Sesamum indicum] |
| 14 |
Hb_000085_430 |
0.0392920744 |
- |
- |
- |
| 15 |
Hb_000092_110 |
0.0392920744 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_000093_120 |
0.0392920744 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_000120_110 |
0.0392920744 |
- |
- |
1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase, putative [Ricinus communis] |
| 18 |
Hb_000125_160 |
0.0392920744 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_000127_060 |
0.0392920744 |
- |
- |
- |
| 20 |
Hb_000128_020 |
0.0392920744 |
- |
- |
- |