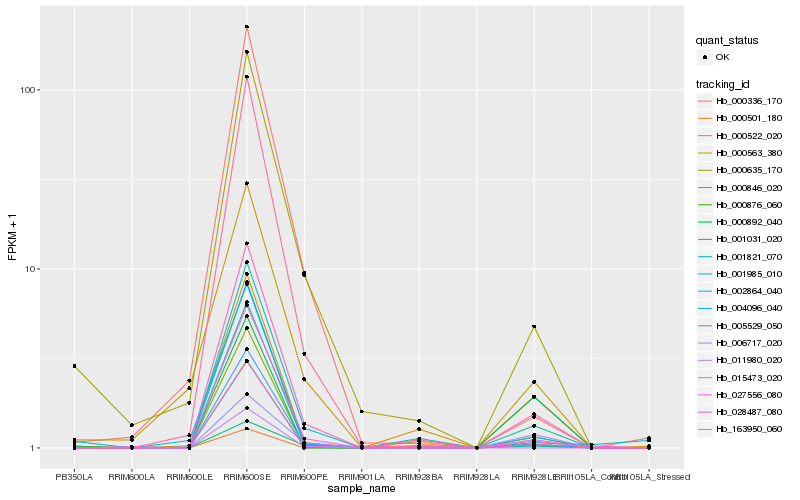
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001821_070 |
0.0 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |
| 2 |
Hb_000846_020 |
0.0642542646 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_015473_020 |
0.0862981672 |
- |
- |
hypothetical protein B456_001G090400 [Gossypium raimondii] |
| 4 |
Hb_028487_080 |
0.0921564545 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_005529_050 |
0.1089438251 |
- |
- |
PREDICTED: cytochrome P450 94B3-like [Nelumbo nucifera] |
| 6 |
Hb_000876_060 |
0.1180490914 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000501_180 |
0.1385697794 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 8 |
Hb_004096_040 |
0.1405937283 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_027556_080 |
0.1446406578 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g24130 [Jatropha curcas] |
| 10 |
Hb_000522_020 |
0.1503352181 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 11 |
Hb_000635_170 |
0.1644863792 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_002864_040 |
0.1654581955 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 13 |
Hb_001985_010 |
0.1669014791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_163950_060 |
0.1678315255 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 15 |
Hb_000892_040 |
0.1715264655 |
- |
- |
- |
| 16 |
Hb_011980_020 |
0.1749877515 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_001031_020 |
0.17631464 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 18 |
Hb_000563_380 |
0.1792926609 |
- |
- |
hypothetical protein CICLE_v10007206mg [Citrus clementina] |
| 19 |
Hb_006717_020 |
0.1808734692 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 20 |
Hb_000336_170 |
0.1811191642 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |