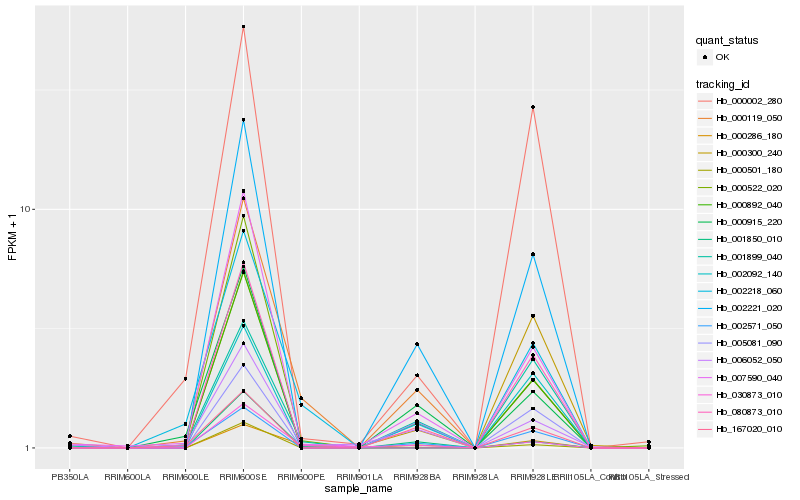
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002221_020 |
0.0 |
- |
- |
hypothetical protein B456_008G083700 [Gossypium raimondii] |
| 2 |
Hb_007590_040 |
0.1189389219 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like [Jatropha curcas] |
| 3 |
Hb_002571_050 |
0.1202284386 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 4 |
Hb_000915_220 |
0.1292436701 |
- |
- |
Lactoylglutathione lyase / glyoxalase I family protein [Theobroma cacao] |
| 5 |
Hb_000501_180 |
0.138973417 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 6 |
Hb_005081_090 |
0.1431292404 |
- |
- |
- |
| 7 |
Hb_000522_020 |
0.1529038503 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 8 |
Hb_001850_010 |
0.1639681066 |
- |
- |
PREDICTED: cytochrome P450 71B34-like [Jatropha curcas] |
| 9 |
Hb_000892_040 |
0.1727696772 |
- |
- |
- |
| 10 |
Hb_080873_010 |
0.1742852869 |
- |
- |
- |
| 11 |
Hb_000002_280 |
0.1783643233 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 12 |
Hb_000300_240 |
0.181243026 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 13 |
Hb_006052_050 |
0.1821645489 |
- |
- |
hypothetical protein EUGRSUZ_A00568 [Eucalyptus grandis] |
| 14 |
Hb_001899_040 |
0.1917059871 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 15 |
Hb_030873_010 |
0.1937209015 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 isoform X2 [Cucumis sativus] |
| 16 |
Hb_167020_010 |
0.2001484315 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_002092_140 |
0.2091687998 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 18 |
Hb_002218_060 |
0.2095214693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000286_180 |
0.2157373024 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 20 |
Hb_000119_050 |
0.2163675085 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |