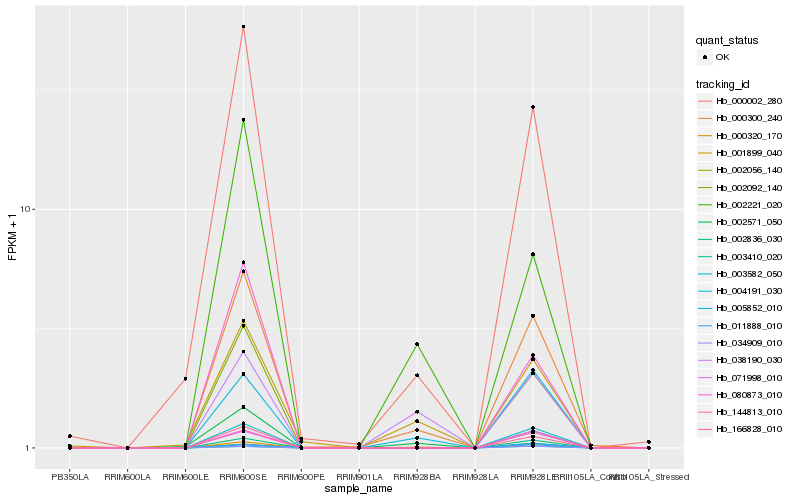
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_240 |
0.0 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 2 |
Hb_000002_280 |
0.1132512037 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 3 |
Hb_166828_010 |
0.1292855611 |
- |
- |
- |
| 4 |
Hb_002092_140 |
0.133344825 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 5 |
Hb_000320_170 |
0.1364793269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002836_030 |
0.1428711469 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 7 |
Hb_003582_050 |
0.1446306554 |
- |
- |
hypothetical protein POPTR_0008s13950g [Populus trichocarpa] |
| 8 |
Hb_004191_030 |
0.1471557682 |
- |
- |
hypothetical protein POPTR_0013s13140g [Populus trichocarpa] |
| 9 |
Hb_001899_040 |
0.1481357342 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 10 |
Hb_144813_010 |
0.1513266557 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 11 |
Hb_002571_050 |
0.1587393019 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 12 |
Hb_011888_010 |
0.1710683279 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 13 |
Hb_071998_010 |
0.1737201432 |
- |
- |
PREDICTED: uncharacterized protein LOC105800957 [Gossypium raimondii] |
| 14 |
Hb_080873_010 |
0.1811508911 |
- |
- |
- |
| 15 |
Hb_002221_020 |
0.181243026 |
- |
- |
hypothetical protein B456_008G083700 [Gossypium raimondii] |
| 16 |
Hb_002056_140 |
0.1837509968 |
- |
- |
- |
| 17 |
Hb_005852_010 |
0.1931161758 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 18 |
Hb_038190_030 |
0.1934053837 |
- |
- |
- |
| 19 |
Hb_034909_010 |
0.1937521051 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_003410_020 |
0.1938389667 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |