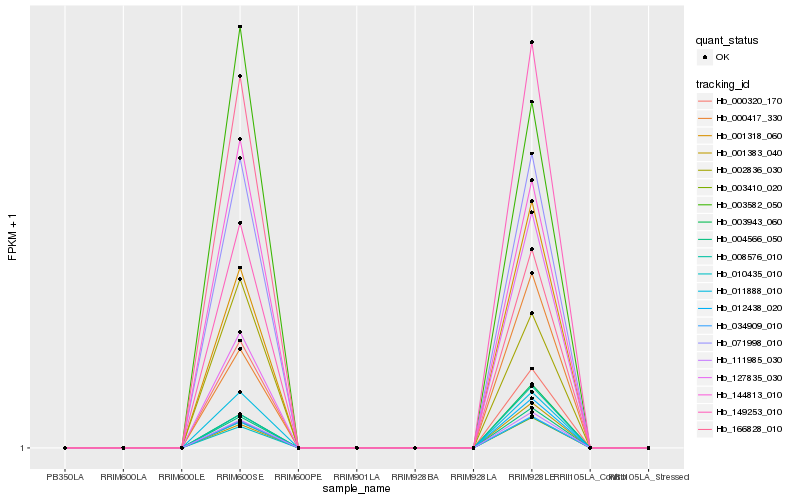
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071998_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105800957 [Gossypium raimondii] |
| 2 |
Hb_011888_010 |
0.0039709659 |
- |
- |
Receptor like protein 46, putative [Theobroma cacao] |
| 3 |
Hb_034909_010 |
0.0281818858 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_003410_020 |
0.0282986311 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_144813_010 |
0.036644084 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 6 |
Hb_004566_050 |
0.0494204194 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_003582_050 |
0.0498336323 |
- |
- |
hypothetical protein POPTR_0008s13950g [Populus trichocarpa] |
| 8 |
Hb_111985_030 |
0.0498355672 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 9 |
Hb_002836_030 |
0.0536172398 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 10 |
Hb_001318_060 |
0.0664501906 |
- |
- |
contains similarity to reverse transcriptase-like proteins [Arabidopsis thaliana] |
| 11 |
Hb_000320_170 |
0.0691723528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010435_010 |
0.0750740207 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_000417_330 |
0.1195245859 |
- |
- |
- |
| 14 |
Hb_008576_010 |
0.1257544601 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_149253_010 |
0.1299668365 |
- |
- |
2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Gossypium arboreum] |
| 16 |
Hb_003943_060 |
0.1307485344 |
- |
- |
PREDICTED: uncharacterized protein LOC105636357 [Jatropha curcas] |
| 17 |
Hb_012438_020 |
0.1320690912 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_001383_040 |
0.1324742248 |
- |
- |
PREDICTED: uncharacterized protein LOC105627956 [Jatropha curcas] |
| 19 |
Hb_166828_010 |
0.1441979951 |
- |
- |
- |
| 20 |
Hb_127835_030 |
0.1502702172 |
- |
- |
- |