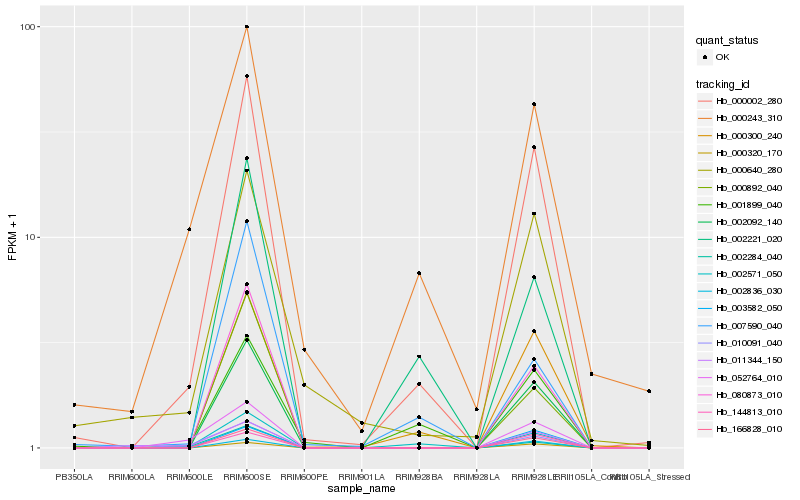
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000002_280 |
0.0 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 2 |
Hb_000300_240 |
0.1132512037 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 3 |
Hb_002092_140 |
0.1163063087 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 4 |
Hb_166828_010 |
0.11852582 |
- |
- |
- |
| 5 |
Hb_002571_050 |
0.1353658901 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 6 |
Hb_080873_010 |
0.1414719896 |
- |
- |
- |
| 7 |
Hb_010091_040 |
0.1488874512 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 8 |
Hb_000320_170 |
0.1524260618 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001899_040 |
0.1616328618 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 10 |
Hb_002836_030 |
0.1628330102 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 11 |
Hb_002284_040 |
0.1629759278 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_003582_050 |
0.1654790534 |
- |
- |
hypothetical protein POPTR_0008s13950g [Populus trichocarpa] |
| 13 |
Hb_144813_010 |
0.1750062128 |
- |
- |
PREDICTED: uncharacterized protein LOC105781565 [Gossypium raimondii] |
| 14 |
Hb_000640_280 |
0.1760506756 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |
| 15 |
Hb_002221_020 |
0.1783643233 |
- |
- |
hypothetical protein B456_008G083700 [Gossypium raimondii] |
| 16 |
Hb_052764_010 |
0.178435514 |
- |
- |
hypothetical protein POPTR_0011s04320g [Populus trichocarpa] |
| 17 |
Hb_000892_040 |
0.1791075981 |
- |
- |
- |
| 18 |
Hb_000243_310 |
0.1818616856 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 19 |
Hb_011344_150 |
0.1963129671 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 20 |
Hb_007590_040 |
0.1994160005 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like [Jatropha curcas] |