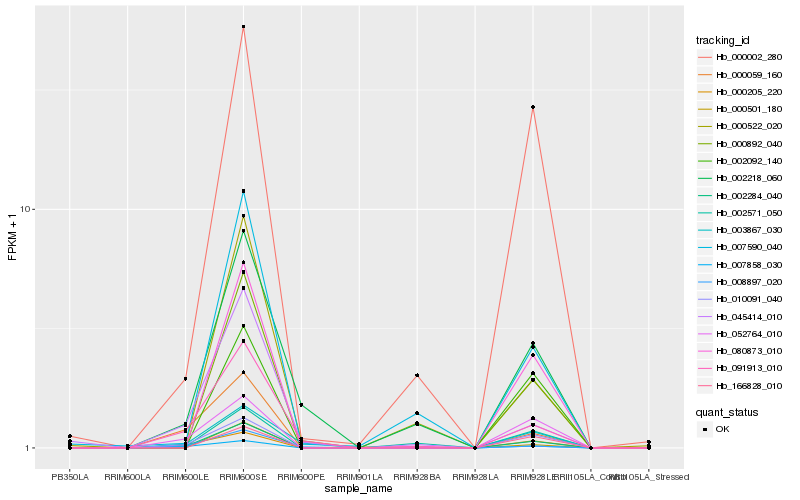
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_040 |
0.0 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_010091_040 |
0.1011530279 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 3 |
Hb_000892_040 |
0.1433290651 |
- |
- |
- |
| 4 |
Hb_080873_010 |
0.1452872766 |
- |
- |
- |
| 5 |
Hb_007858_030 |
0.1469274292 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |
| 6 |
Hb_052764_010 |
0.1548247186 |
- |
- |
hypothetical protein POPTR_0011s04320g [Populus trichocarpa] |
| 7 |
Hb_000205_220 |
0.1570433035 |
- |
- |
PREDICTED: uncharacterized protein LOC105647760 [Jatropha curcas] |
| 8 |
Hb_000002_280 |
0.1629759278 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 9 |
Hb_091913_010 |
0.1753213938 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 10 |
Hb_000059_160 |
0.184088783 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 11 |
Hb_002092_140 |
0.1862107495 |
- |
- |
hypothetical protein EUGRSUZ_B00708, partial [Eucalyptus grandis] |
| 12 |
Hb_003867_030 |
0.1914438332 |
- |
- |
- |
| 13 |
Hb_007590_040 |
0.193111648 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like [Jatropha curcas] |
| 14 |
Hb_166828_010 |
0.197062784 |
- |
- |
- |
| 15 |
Hb_002218_060 |
0.2021478635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002571_050 |
0.2023136057 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 17 |
Hb_000501_180 |
0.2023379232 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 18 |
Hb_008897_020 |
0.2133263202 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_000522_020 |
0.2138965406 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 20 |
Hb_045414_010 |
0.2144634526 |
- |
- |
unnamed protein product [Vitis vinifera] |