| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
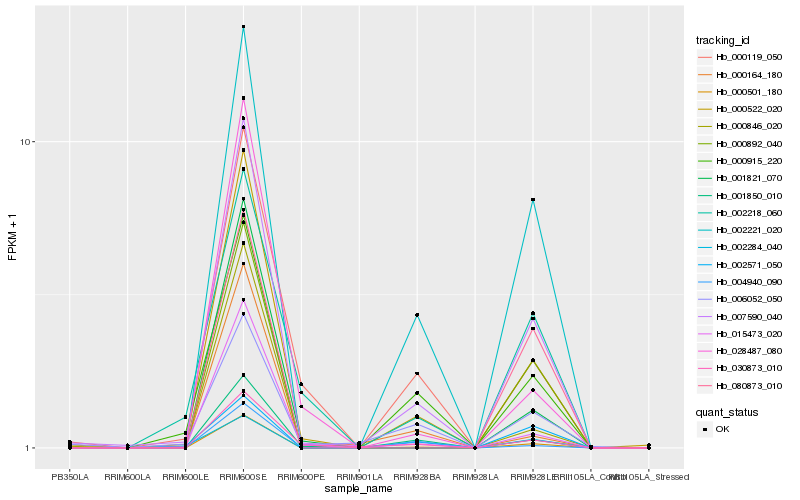
Hb_007590_040 |
0.0 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like [Jatropha curcas] |
| 2 |
Hb_000522_020 |
0.069743188 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 3 |
Hb_000501_180 |
0.0916152769 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 4 |
Hb_002221_020 |
0.1189389219 |
- |
- |
hypothetical protein B456_008G083700 [Gossypium raimondii] |
| 5 |
Hb_001850_010 |
0.1369612328 |
- |
- |
PREDICTED: cytochrome P450 71B34-like [Jatropha curcas] |
| 6 |
Hb_000915_220 |
0.137346756 |
- |
- |
Lactoylglutathione lyase / glyoxalase I family protein [Theobroma cacao] |
| 7 |
Hb_015473_020 |
0.1474552439 |
- |
- |
hypothetical protein B456_001G090400 [Gossypium raimondii] |
| 8 |
Hb_030873_010 |
0.1515737858 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 isoform X2 [Cucumis sativus] |
| 9 |
Hb_000892_040 |
0.1545764783 |
- |
- |
- |
| 10 |
Hb_006052_050 |
0.1563805965 |
- |
- |
hypothetical protein EUGRSUZ_A00568 [Eucalyptus grandis] |
| 11 |
Hb_000119_050 |
0.1675711141 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 12 |
Hb_080873_010 |
0.1800159792 |
- |
- |
- |
| 13 |
Hb_001821_070 |
0.1812781057 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |
| 14 |
Hb_002571_050 |
0.1831209978 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 15 |
Hb_000164_180 |
0.184832561 |
- |
- |
(+)-delta-cadinene synthase isozyme A, putative [Ricinus communis] |
| 16 |
Hb_028487_080 |
0.188037514 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 17 |
Hb_002218_060 |
0.1887357958 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002284_040 |
0.193111648 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000846_020 |
0.1961525694 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_004940_090 |
0.1970734686 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |