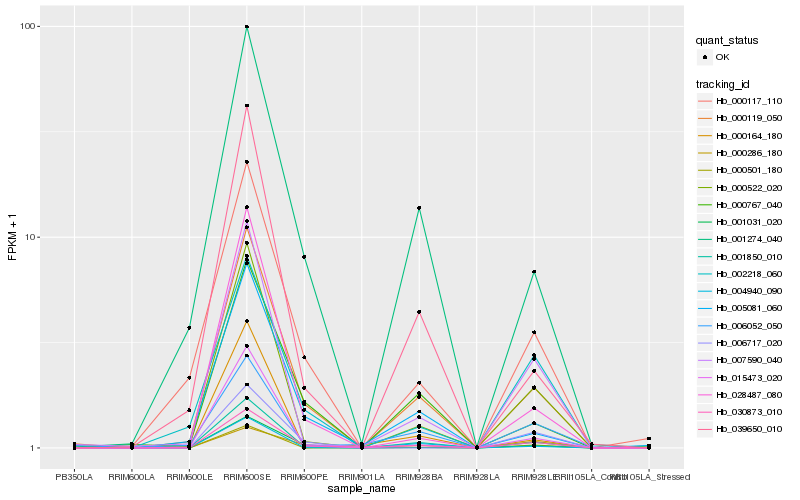
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030873_010 |
0.0 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 isoform X2 [Cucumis sativus] |
| 2 |
Hb_001850_010 |
0.1214244237 |
- |
- |
PREDICTED: cytochrome P450 71B34-like [Jatropha curcas] |
| 3 |
Hb_000119_050 |
0.1230126837 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 4 |
Hb_000522_020 |
0.1408594251 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 5 |
Hb_007590_040 |
0.1515737858 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like [Jatropha curcas] |
| 6 |
Hb_015473_020 |
0.1567059543 |
- |
- |
hypothetical protein B456_001G090400 [Gossypium raimondii] |
| 7 |
Hb_028487_080 |
0.1671480698 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001031_020 |
0.1700058289 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 9 |
Hb_000501_180 |
0.1710651644 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g67520 [Jatropha curcas] |
| 10 |
Hb_006052_050 |
0.1715312293 |
- |
- |
hypothetical protein EUGRSUZ_A00568 [Eucalyptus grandis] |
| 11 |
Hb_005081_060 |
0.1717835938 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 12 |
Hb_039650_010 |
0.1807341959 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 13 |
Hb_000117_110 |
0.1814673118 |
- |
- |
transferase, putative [Ricinus communis] |
| 14 |
Hb_000164_180 |
0.1840063139 |
- |
- |
(+)-delta-cadinene synthase isozyme A, putative [Ricinus communis] |
| 15 |
Hb_006717_020 |
0.1842573886 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 16 |
Hb_000767_040 |
0.1844543125 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_002218_060 |
0.1851052444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001274_040 |
0.1866913408 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 19 |
Hb_004940_090 |
0.1873765307 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 20 |
Hb_000286_180 |
0.1917428126 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |