| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
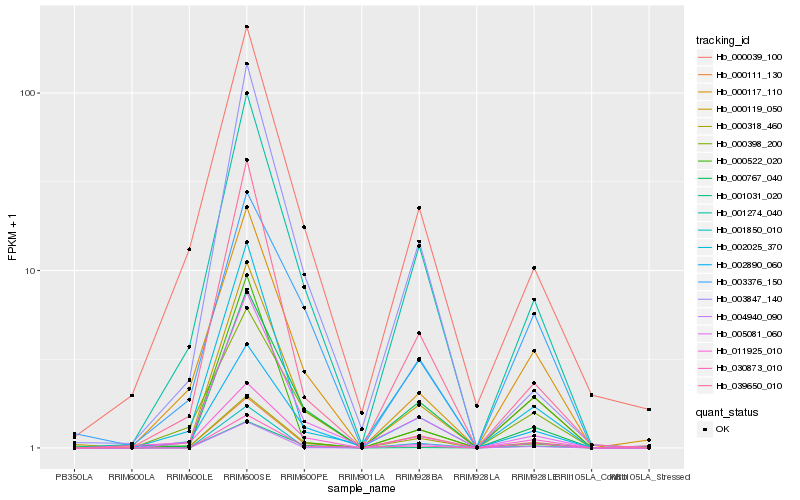
Hb_000119_050 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 2 |
Hb_001850_010 |
0.1102799997 |
- |
- |
PREDICTED: cytochrome P450 71B34-like [Jatropha curcas] |
| 3 |
Hb_001274_040 |
0.1103424805 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 4 |
Hb_000767_040 |
0.1209320174 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_030873_010 |
0.1230126837 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 isoform X2 [Cucumis sativus] |
| 6 |
Hb_000111_130 |
0.1285182278 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 7 |
Hb_005081_060 |
0.1290184471 |
- |
- |
hypothetical protein EUGRSUZ_C00833 [Eucalyptus grandis] |
| 8 |
Hb_039650_010 |
0.129536465 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |
| 9 |
Hb_000117_110 |
0.1347875385 |
- |
- |
transferase, putative [Ricinus communis] |
| 10 |
Hb_000522_020 |
0.1411221934 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 11 |
Hb_001031_020 |
0.141403001 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 12 |
Hb_002890_060 |
0.1416692715 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 13 |
Hb_000398_200 |
0.1468562595 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 14 |
Hb_000318_460 |
0.1492546814 |
- |
- |
hypothetical protein CICLE_v10028333mg [Citrus clementina] |
| 15 |
Hb_002025_370 |
0.1560568297 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 16 |
Hb_000039_100 |
0.1595808896 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 17 |
Hb_003376_150 |
0.1618454384 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 18 |
Hb_004940_090 |
0.1639565764 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 19 |
Hb_011925_010 |
0.1646184263 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003847_140 |
0.1670500249 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |