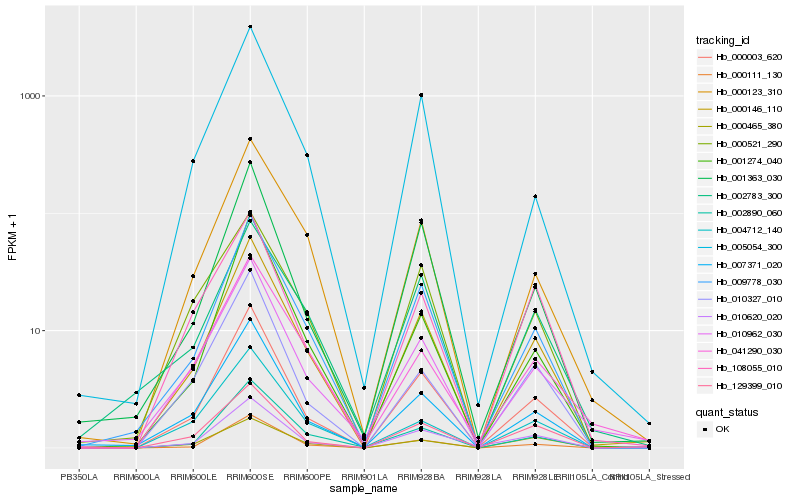
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000146_110 |
0.0 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 2 |
Hb_000003_620 |
0.0587718399 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 3 |
Hb_009778_030 |
0.0610917365 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 4 |
Hb_002890_060 |
0.0905107589 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 5 |
Hb_000111_130 |
0.0914312314 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 6 |
Hb_000123_310 |
0.0933945634 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 7 |
Hb_007371_020 |
0.0954584125 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_010962_030 |
0.1006892651 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_010620_020 |
0.1083896799 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_129399_010 |
0.1093708968 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_001274_040 |
0.1136293697 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 12 |
Hb_001363_030 |
0.1139894687 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 13 |
Hb_004712_140 |
0.1156675583 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 14 |
Hb_010327_010 |
0.1197959344 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_041290_030 |
0.120343504 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 16 |
Hb_005054_300 |
0.121162807 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 17 |
Hb_002783_300 |
0.1237960679 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000521_290 |
0.1244839317 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 19 |
Hb_108055_010 |
0.1260791633 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000465_380 |
0.126449181 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |