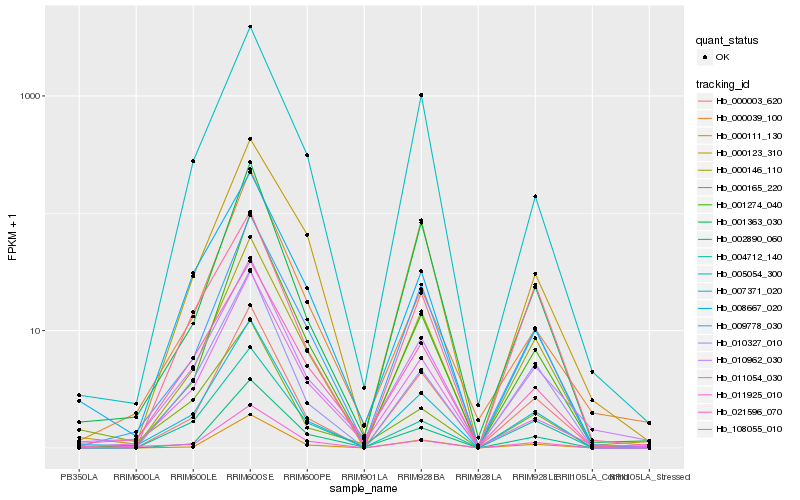
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007371_020 |
0.0 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_010962_030 |
0.0805225974 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_000003_620 |
0.0906644301 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 4 |
Hb_021596_070 |
0.0938361686 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 5 |
Hb_000146_110 |
0.0954584125 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 6 |
Hb_009778_030 |
0.0960464203 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 7 |
Hb_010327_010 |
0.0965353398 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 8 |
Hb_008667_020 |
0.1085210809 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_004712_140 |
0.1094353232 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 10 |
Hb_001274_040 |
0.1116922377 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 11 |
Hb_011925_010 |
0.1144194971 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011054_030 |
0.1183456572 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 13 |
Hb_000111_130 |
0.118364351 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 14 |
Hb_002890_060 |
0.1193252051 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 15 |
Hb_005054_300 |
0.1219311453 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 16 |
Hb_000123_310 |
0.1224581783 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 17 |
Hb_001363_030 |
0.1235428605 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 18 |
Hb_000039_100 |
0.12918471 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 19 |
Hb_000165_220 |
0.1317285244 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 20 |
Hb_108055_010 |
0.1380508446 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |