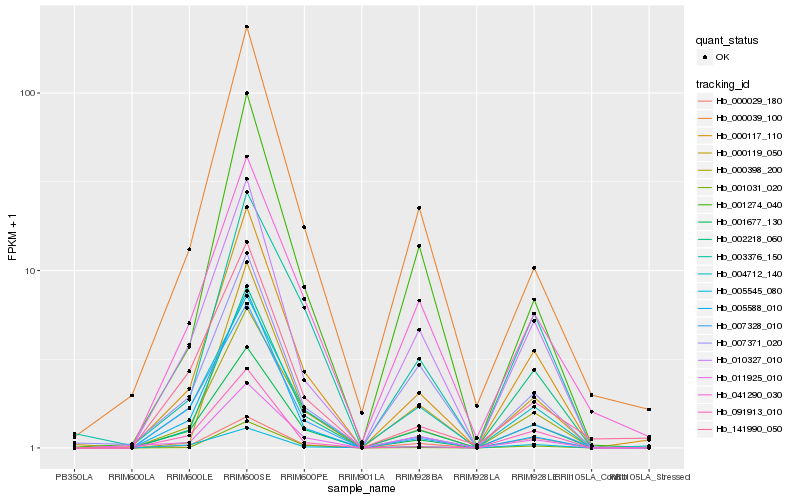
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000117_110 |
0.0 |
- |
- |
transferase, putative [Ricinus communis] |
| 2 |
Hb_000398_200 |
0.0683113195 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 3 |
Hb_001677_130 |
0.0752129866 |
- |
- |
hypothetical protein POPTR_0081s002101g, partial [Populus trichocarpa] |
| 4 |
Hb_004712_140 |
0.1126829735 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 5 |
Hb_010327_010 |
0.1192800688 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 6 |
Hb_002218_060 |
0.1232757634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_091913_010 |
0.1337534354 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 8 |
Hb_007328_010 |
0.1343942 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_000119_050 |
0.1347875385 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 10 |
Hb_005545_080 |
0.1399760617 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 11 |
Hb_000039_100 |
0.1413769705 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 12 |
Hb_011925_010 |
0.1413813372 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001274_040 |
0.1436782411 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 14 |
Hb_001031_020 |
0.1437086175 |
- |
- |
hypothetical protein VITISV_000631 [Vitis vinifera] |
| 15 |
Hb_141990_050 |
0.1456407667 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 16 |
Hb_003376_150 |
0.1460985637 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 17 |
Hb_041290_030 |
0.1494615294 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 18 |
Hb_000029_180 |
0.1521912412 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 19 |
Hb_005588_010 |
0.1601620919 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 20 |
Hb_007371_020 |
0.1612195072 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |