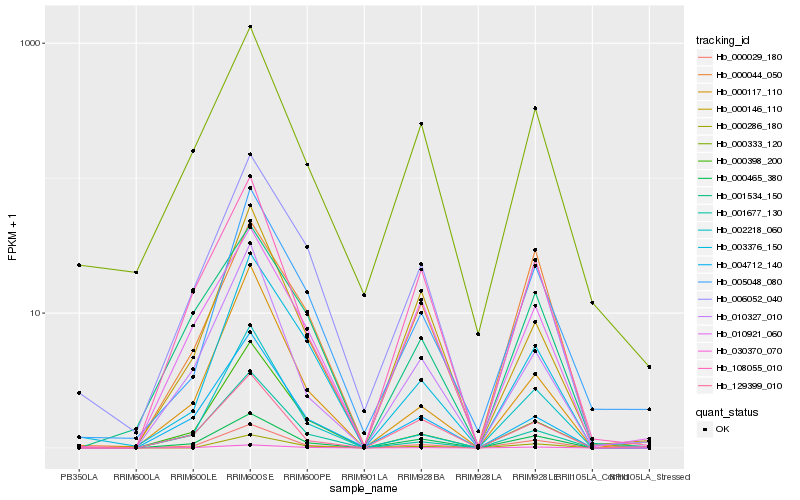
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000029_180 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 2 |
Hb_000465_380 |
0.077006035 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_030370_070 |
0.0875289215 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_002218_060 |
0.1100339798 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_108055_010 |
0.1110698982 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_010327_010 |
0.1248308142 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_129399_010 |
0.1354638057 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_005048_080 |
0.1416992159 |
- |
- |
PREDICTED: PAP-specific phosphatase HAL2-like [Jatropha curcas] |
| 9 |
Hb_004712_140 |
0.1505140864 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 10 |
Hb_001677_130 |
0.1506670868 |
- |
- |
hypothetical protein POPTR_0081s002101g, partial [Populus trichocarpa] |
| 11 |
Hb_000117_110 |
0.1521912412 |
- |
- |
transferase, putative [Ricinus communis] |
| 12 |
Hb_003376_150 |
0.1526853512 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 13 |
Hb_000286_180 |
0.153222794 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 14 |
Hb_000333_120 |
0.1540078822 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 15 |
Hb_000146_110 |
0.1540533407 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 16 |
Hb_000398_200 |
0.1545790636 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 17 |
Hb_000044_050 |
0.1556183498 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001534_150 |
0.1593395905 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 19 |
Hb_010921_060 |
0.1595747197 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_006052_040 |
0.1642984706 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |