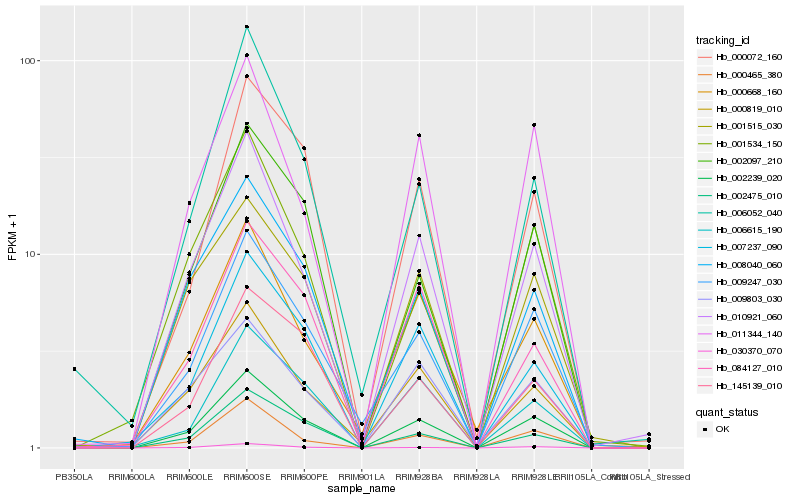
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002239_020 |
0.0 |
- |
- |
MLO, putative [Theobroma cacao] |
| 2 |
Hb_010921_060 |
0.0863531442 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_009247_030 |
0.0941536206 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 4 |
Hb_002097_210 |
0.1034563226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007237_090 |
0.1047107414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000465_380 |
0.1069713462 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_008040_060 |
0.1109647966 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 8 |
Hb_006615_190 |
0.1152552604 |
- |
- |
UDP-glycosyltransferase 1 [Linum usitatissimum] |
| 9 |
Hb_000819_010 |
0.1165865076 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 10 |
Hb_000072_160 |
0.1179265874 |
- |
- |
proline oxidase [Manihot esculenta] |
| 11 |
Hb_011344_140 |
0.1180153965 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 12 |
Hb_001515_030 |
0.1189201657 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_030370_070 |
0.1230126997 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000668_160 |
0.1252089283 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 15 |
Hb_001534_150 |
0.1256118358 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 16 |
Hb_145139_010 |
0.1270426548 |
- |
- |
PREDICTED: sugar transporter ERD6-like 5 isoform X1 [Vitis vinifera] |
| 17 |
Hb_084127_010 |
0.1278675932 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_009803_030 |
0.1279978569 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 19 |
Hb_002475_010 |
0.1294821829 |
- |
- |
hypothetical protein JCGZ_22530 [Jatropha curcas] |
| 20 |
Hb_006052_040 |
0.1311202254 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |