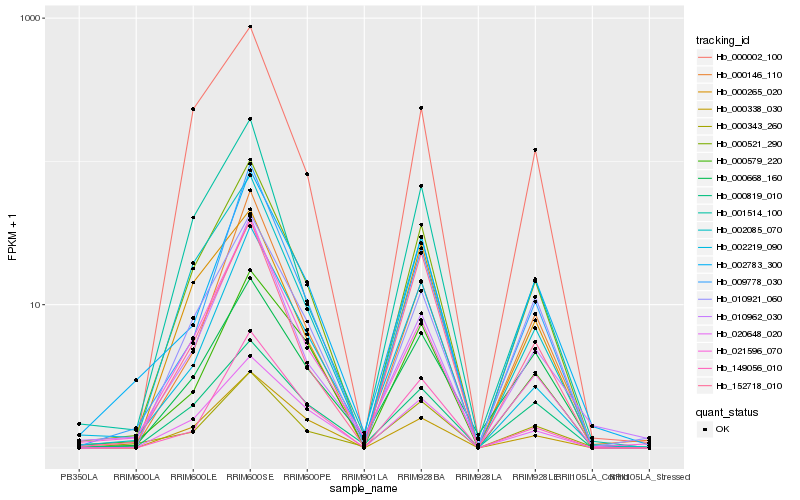
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_290 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 2 |
Hb_002085_070 |
0.0822591804 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000002_100 |
0.0923528582 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 4 |
Hb_020648_020 |
0.1013514138 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor pho1, putative [Ricinus communis] |
| 5 |
Hb_010921_060 |
0.1050204118 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000338_030 |
0.1120476494 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_21254 [Jatropha curcas] |
| 7 |
Hb_001514_100 |
0.113103214 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 8 |
Hb_000819_010 |
0.113140013 |
- |
- |
Receptor-like protein kinase 1, putative [Theobroma cacao] |
| 9 |
Hb_000265_020 |
0.1144466657 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 10 |
Hb_000343_260 |
0.1223482287 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 11 |
Hb_009778_030 |
0.1237051651 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 12 |
Hb_002783_300 |
0.1240476908 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000146_110 |
0.1244839317 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 14 |
Hb_000579_220 |
0.125838859 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000668_160 |
0.1261522547 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 16 |
Hb_152718_010 |
0.1275728839 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 17 |
Hb_021596_070 |
0.1291215711 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_002219_090 |
0.1344082912 |
- |
- |
protein kinase atmrk1, putative [Ricinus communis] |
| 19 |
Hb_149056_010 |
0.1364860313 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |
| 20 |
Hb_010962_030 |
0.138459615 |
- |
- |
kinase, putative [Ricinus communis] |