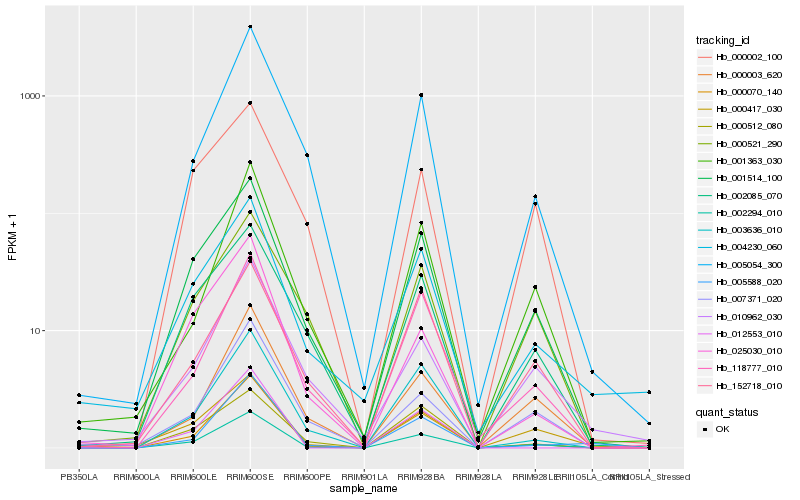
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002294_010 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 2 |
Hb_000070_140 |
0.0834859417 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_001514_100 |
0.0973154132 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 4 |
Hb_000417_030 |
0.1325169779 |
transcription factor |
TF Family: G2-like |
PREDICTED: putative two-component response regulator ARR19 [Jatropha curcas] |
| 5 |
Hb_118777_010 |
0.1329441858 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001363_030 |
0.1347611645 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 7 |
Hb_000003_620 |
0.136453015 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 8 |
Hb_003636_010 |
0.1366332945 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 9 |
Hb_005054_300 |
0.1401343266 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 10 |
Hb_007371_020 |
0.1507733601 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002085_070 |
0.1533543348 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_005588_020 |
0.1560578506 |
- |
- |
PREDICTED: patatin-like protein 2 [Vitis vinifera] |
| 13 |
Hb_152718_010 |
0.1570294001 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 14 |
Hb_004230_060 |
0.1571428321 |
- |
- |
PREDICTED: ferritin, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_000512_080 |
0.1627465937 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_025030_010 |
0.163509782 |
- |
- |
- |
| 17 |
Hb_000521_290 |
0.1640336678 |
- |
- |
hypothetical protein POPTR_0001s23530g [Populus trichocarpa] |
| 18 |
Hb_012553_010 |
0.1656813143 |
- |
- |
hypothetical protein JCGZ_10552 [Jatropha curcas] |
| 19 |
Hb_010962_030 |
0.1662262605 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_000002_100 |
0.1672193415 |
- |
- |
phosphate transporter [Manihot esculenta] |