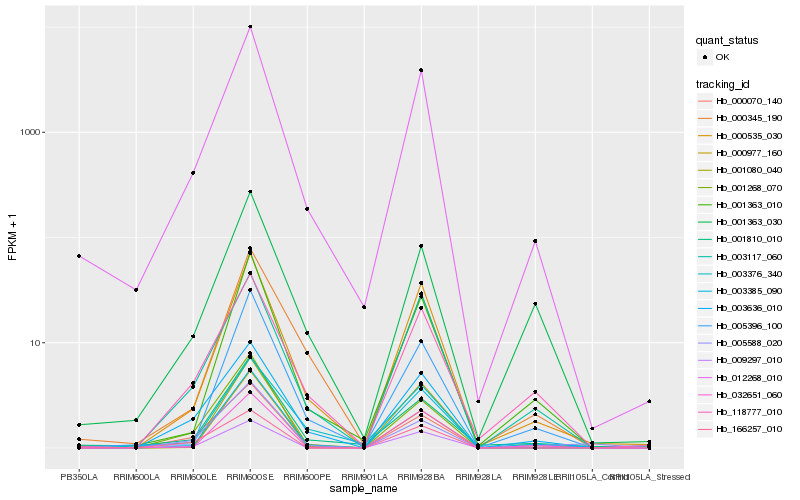
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012268_010 |
0.0 |
- |
- |
glutamic acid-rich protein [Manihot esculenta] |
| 2 |
Hb_000535_030 |
0.1010437342 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 3 |
Hb_000070_140 |
0.1052725613 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_001363_010 |
0.1075233957 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 5 |
Hb_003636_010 |
0.1118759843 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 6 |
Hb_001810_010 |
0.1137816909 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 7 |
Hb_003117_060 |
0.1212251749 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 8 |
Hb_005396_100 |
0.1239417929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000345_190 |
0.1271097215 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 10 |
Hb_003385_090 |
0.1272305515 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 11 |
Hb_118777_010 |
0.1296721256 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_009297_010 |
0.1303515411 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_032651_060 |
0.1313090763 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000977_160 |
0.1348043305 |
- |
- |
Serine-threonine protein kinase, plant-type, putative [Theobroma cacao] |
| 15 |
Hb_001268_070 |
0.1375899899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_166257_010 |
0.1435990301 |
- |
- |
hypothetical protein JCGZ_21808 [Jatropha curcas] |
| 17 |
Hb_005588_020 |
0.1451087677 |
- |
- |
PREDICTED: patatin-like protein 2 [Vitis vinifera] |
| 18 |
Hb_003376_340 |
0.1453601823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001080_040 |
0.1469272347 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_001363_030 |
0.1496065255 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |