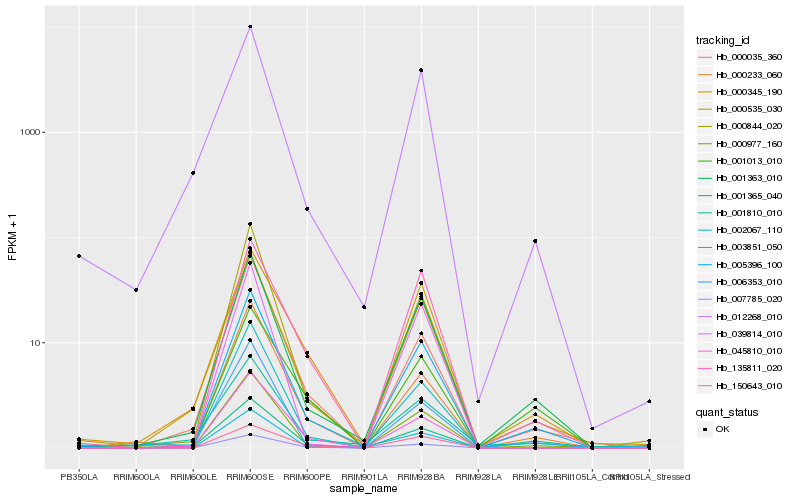
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005396_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001363_010 |
0.0614799031 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 3 |
Hb_000977_160 |
0.0741887748 |
- |
- |
Serine-threonine protein kinase, plant-type, putative [Theobroma cacao] |
| 4 |
Hb_000233_060 |
0.1034724348 |
- |
- |
hypothetical protein JCGZ_14793 [Jatropha curcas] |
| 5 |
Hb_007785_020 |
0.1048429307 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_002067_110 |
0.1052612323 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Populus euphratica] |
| 7 |
Hb_000345_190 |
0.1143669049 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 8 |
Hb_045810_010 |
0.1176151186 |
- |
- |
PREDICTED: laccase-12-like [Jatropha curcas] |
| 9 |
Hb_000035_360 |
0.1180287744 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 10 |
Hb_000844_020 |
0.1192294823 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 11 |
Hb_135811_020 |
0.1199976958 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 12 |
Hb_012268_010 |
0.1239417929 |
- |
- |
glutamic acid-rich protein [Manihot esculenta] |
| 13 |
Hb_000535_030 |
0.1263250979 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 14 |
Hb_006353_010 |
0.1276838656 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 15 |
Hb_003851_050 |
0.1288521264 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 16 |
Hb_001365_040 |
0.1314237743 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X2 [Populus euphratica] |
| 17 |
Hb_001013_010 |
0.1330740544 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_150643_010 |
0.1345914984 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 19 |
Hb_001810_010 |
0.1374710974 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 20 |
Hb_039814_010 |
0.1390687601 |
- |
- |
hypothetical protein JCGZ_05023 [Jatropha curcas] |