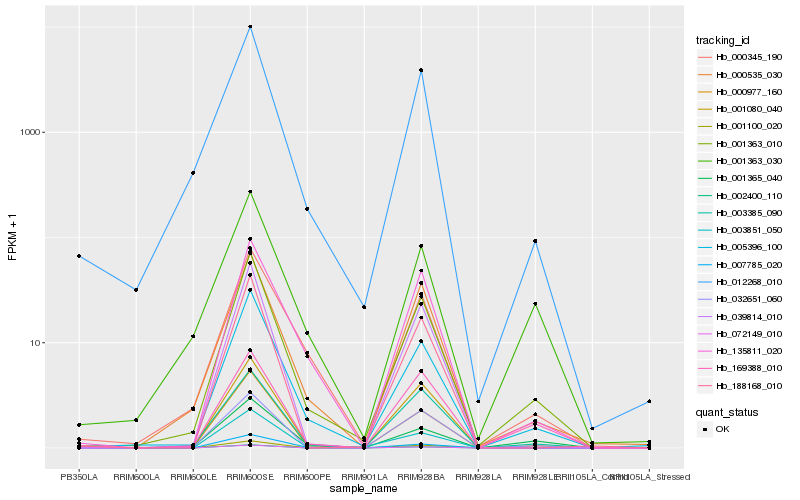
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001363_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 2 |
Hb_005396_100 |
0.0614799031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000977_160 |
0.0879006286 |
- |
- |
Serine-threonine protein kinase, plant-type, putative [Theobroma cacao] |
| 4 |
Hb_000535_030 |
0.099909918 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 5 |
Hb_003385_090 |
0.1005273906 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 6 |
Hb_032651_060 |
0.104224839 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_012268_010 |
0.1075233957 |
- |
- |
glutamic acid-rich protein [Manihot esculenta] |
| 8 |
Hb_135811_020 |
0.1209747243 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 9 |
Hb_001080_040 |
0.1244684051 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000345_190 |
0.1248544524 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 11 |
Hb_001365_040 |
0.1249271657 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X2 [Populus euphratica] |
| 12 |
Hb_039814_010 |
0.1290928407 |
- |
- |
hypothetical protein JCGZ_05023 [Jatropha curcas] |
| 13 |
Hb_169388_010 |
0.1341602469 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 14 |
Hb_003851_050 |
0.1346648366 |
- |
- |
oligopeptide transporter, putative [Ricinus communis] |
| 15 |
Hb_188168_010 |
0.1389524035 |
- |
- |
Arylacetamide deacetylase, putative [Ricinus communis] |
| 16 |
Hb_007785_020 |
0.1394273302 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001363_030 |
0.1394789367 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 18 |
Hb_002400_110 |
0.1401209753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_072149_010 |
0.140124827 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 20 |
Hb_001100_020 |
0.1401368452 |
- |
- |
- |