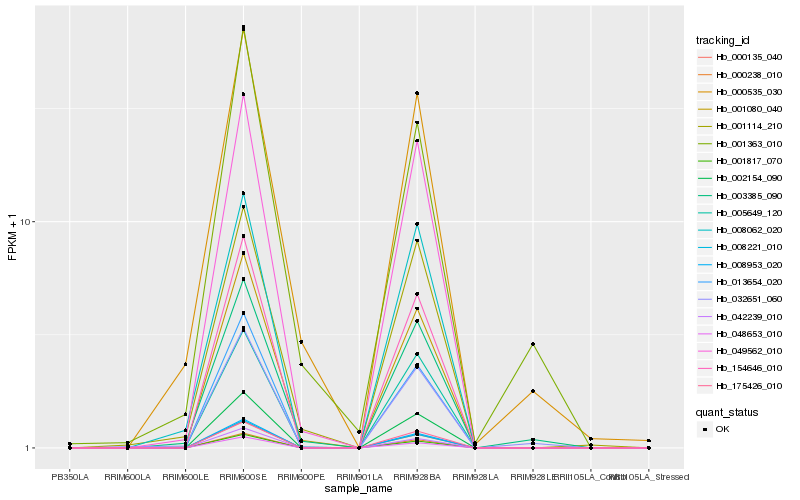
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032651_060 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_003385_090 |
0.0599743428 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 3 |
Hb_000535_030 |
0.0955682894 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 4 |
Hb_049562_010 |
0.0978653495 |
- |
- |
hypothetical protein JCGZ_25442 [Jatropha curcas] |
| 5 |
Hb_001080_040 |
0.1031202891 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_001363_010 |
0.104224839 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 7 |
Hb_005649_120 |
0.1079788518 |
- |
- |
Anthocyanin 5-aromatic acyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000238_010 |
0.1092271796 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_002154_090 |
0.1092621558 |
- |
- |
- |
| 10 |
Hb_001114_210 |
0.1099115271 |
- |
- |
PREDICTED: periaxin [Jatropha curcas] |
| 11 |
Hb_154646_010 |
0.1104417206 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_001817_070 |
0.1109056729 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_175426_010 |
0.1122825207 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 14 |
Hb_000135_040 |
0.1124707955 |
- |
- |
PREDICTED: putative F-box/FBD/LRR-repeat protein At5g56810 [Jatropha curcas] |
| 15 |
Hb_008953_020 |
0.1125164806 |
- |
- |
hypothetical protein EUGRSUZ_C03826 [Eucalyptus grandis] |
| 16 |
Hb_008221_010 |
0.1137901574 |
- |
- |
hypothetical protein JCGZ_10961 [Jatropha curcas] |
| 17 |
Hb_008062_020 |
0.1140449465 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 18 |
Hb_013654_020 |
0.1148560011 |
- |
- |
putative prolin-rich extensin-like receptor protein kinase family protein [Zea mays] |
| 19 |
Hb_048653_010 |
0.114899808 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 20 |
Hb_042239_010 |
0.1152954413 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |