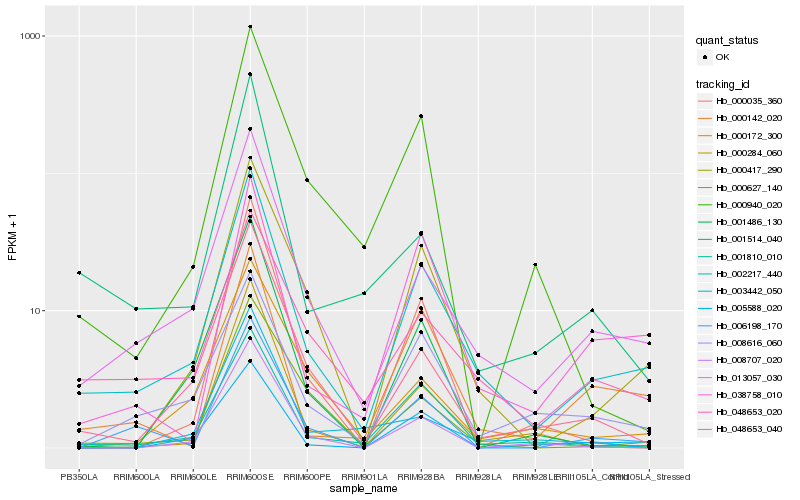
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_440 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_013057_030 |
0.1104496131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001810_010 |
0.134670149 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 4 |
Hb_048653_020 |
0.153049386 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000417_290 |
0.1539054631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008616_060 |
0.1592853118 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 7 |
Hb_048653_040 |
0.1643697122 |
- |
- |
ankyrin repeat family protein [Populus trichocarpa] |
| 8 |
Hb_000142_020 |
0.1711258177 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 9 |
Hb_006198_170 |
0.176432508 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070-like [Jatropha curcas] |
| 10 |
Hb_038758_010 |
0.1768917252 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001514_040 |
0.1886352914 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 12 |
Hb_000172_300 |
0.1902701278 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 13 |
Hb_000627_140 |
0.1911768701 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_001486_130 |
0.192061401 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 15 |
Hb_000940_020 |
0.1958234078 |
- |
- |
hypothetical protein POPTR_0004s17500g [Populus trichocarpa] |
| 16 |
Hb_005588_020 |
0.1971024692 |
- |
- |
PREDICTED: patatin-like protein 2 [Vitis vinifera] |
| 17 |
Hb_000035_360 |
0.1988703341 |
- |
- |
PREDICTED: VQ motif-containing protein 31-like [Jatropha curcas] |
| 18 |
Hb_000284_060 |
0.199655491 |
desease resistance |
Gene Name: NB-ARC |
disease resistance protein (TIR-NBS-LRR class), putative [Medicago truncatula] |
| 19 |
Hb_008707_020 |
0.1997761124 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_003442_050 |
0.2007832462 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |