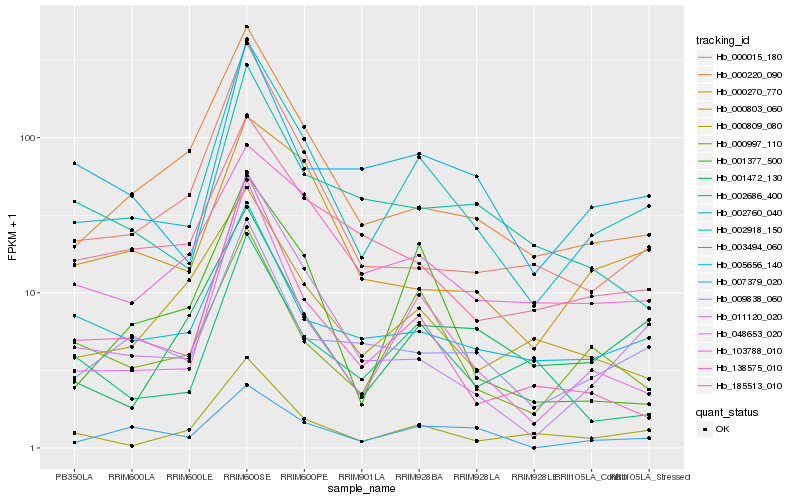
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002918_150 |
0.0 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 2 |
Hb_048653_020 |
0.1271060123 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_011120_020 |
0.1525245757 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000809_080 |
0.1557500629 |
- |
- |
50S ribosomal protein L25, putative [Ricinus communis] |
| 5 |
Hb_000220_090 |
0.1561600581 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 6 |
Hb_138575_010 |
0.1630124947 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 7 |
Hb_009838_060 |
0.1657423417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005656_140 |
0.1715664476 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 9 |
Hb_000997_110 |
0.1719749322 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |
| 10 |
Hb_003494_060 |
0.1735559778 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000015_180 |
0.1744621985 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 12 |
Hb_185513_010 |
0.1769717517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000270_770 |
0.1799405644 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 14 |
Hb_103788_010 |
0.1800525126 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 15 |
Hb_001472_130 |
0.1826140433 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 16 |
Hb_000803_060 |
0.1844047941 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 17 |
Hb_007379_020 |
0.1848365318 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 18 |
Hb_002686_400 |
0.185785995 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 19 |
Hb_002760_040 |
0.1870004081 |
- |
- |
PREDICTED: uncharacterized protein LOC105641234 [Jatropha curcas] |
| 20 |
Hb_001377_500 |
0.1895227375 |
- |
- |
DOF AFFECTING GERMINATION 2 family protein [Populus trichocarpa] |