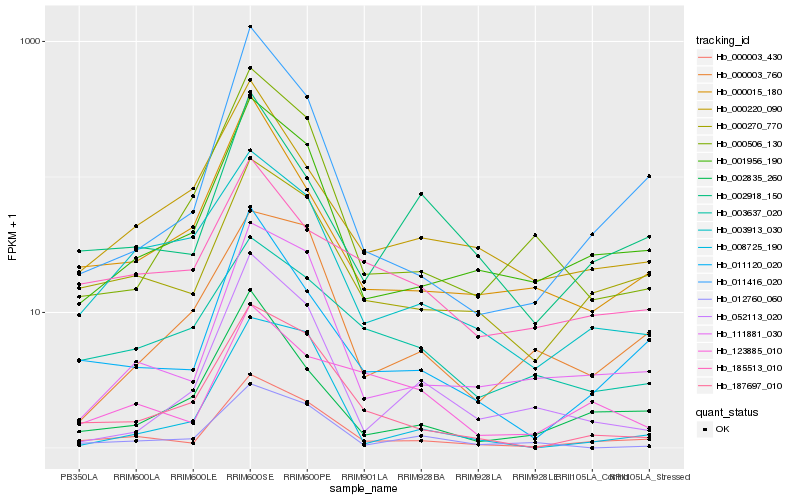
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_430 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 2 |
Hb_111881_030 |
0.1230045937 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 3 |
Hb_001956_190 |
0.1301688644 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 4 |
Hb_000270_770 |
0.1305163243 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 5 |
Hb_011120_020 |
0.1421771853 |
- |
- |
PREDICTED: casein kinase I-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_187697_010 |
0.1501707702 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 7 |
Hb_011416_020 |
0.1605707583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003913_030 |
0.1850215074 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 9 |
Hb_012760_060 |
0.1896560716 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000015_180 |
0.1920300665 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 11 |
Hb_000506_130 |
0.1920796794 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 12 |
Hb_002918_150 |
0.1929215911 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 13 |
Hb_002835_260 |
0.1965197802 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 14 |
Hb_000003_760 |
0.199690089 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 15 |
Hb_008725_190 |
0.2015768859 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 16 |
Hb_000220_090 |
0.2046954909 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHA2A-like [Jatropha curcas] |
| 17 |
Hb_052113_020 |
0.2084713023 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 18 |
Hb_003637_020 |
0.2110259066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_123885_010 |
0.2119268287 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 20 |
Hb_185513_010 |
0.2134493239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |