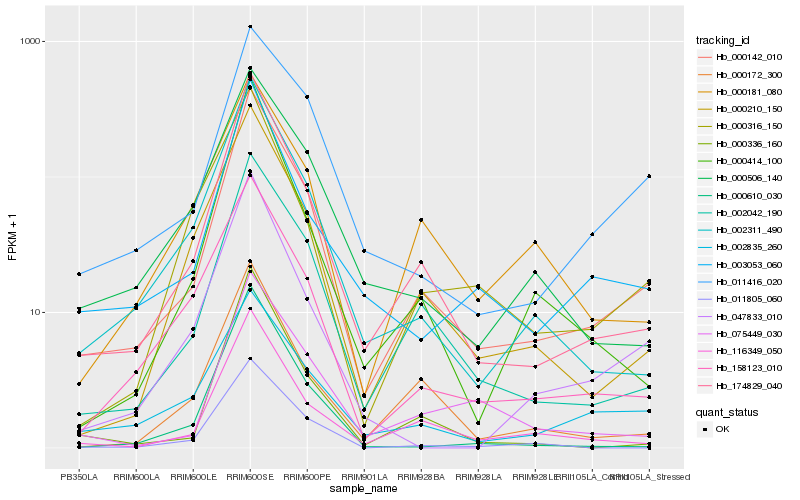
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000142_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 2 |
Hb_174829_040 |
0.08297079 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 3 |
Hb_002042_190 |
0.1029468824 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 4 |
Hb_116349_050 |
0.1252537419 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_002311_490 |
0.1317100904 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 6 |
Hb_011416_020 |
0.1353670182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003053_060 |
0.1366281251 |
- |
- |
PREDICTED: uncharacterized protein LOC105641515 [Jatropha curcas] |
| 8 |
Hb_000210_150 |
0.1386642162 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 9 |
Hb_158123_010 |
0.1433274796 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 10 |
Hb_075449_030 |
0.1493414226 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 11 |
Hb_000336_160 |
0.1500130703 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000172_300 |
0.1502846947 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 13 |
Hb_000414_100 |
0.1507297062 |
- |
- |
PREDICTED: uncharacterized protein LOC105643784 [Jatropha curcas] |
| 14 |
Hb_047833_010 |
0.1539319883 |
- |
- |
Avr9/Cf-9 rapidly elicited protein [Jatropha curcas] |
| 15 |
Hb_000506_140 |
0.1551167445 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 16 |
Hb_002835_260 |
0.1554593945 |
- |
- |
PREDICTED: uncharacterized protein LOC105640313 [Jatropha curcas] |
| 17 |
Hb_011805_060 |
0.1580792367 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 18 |
Hb_000181_080 |
0.1652644859 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 19 |
Hb_000610_030 |
0.1676766782 |
- |
- |
hypothetical protein MIMGU_mgv1a0038491mg, partial [Erythranthe guttata] |
| 20 |
Hb_000316_150 |
0.1698484005 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |