| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
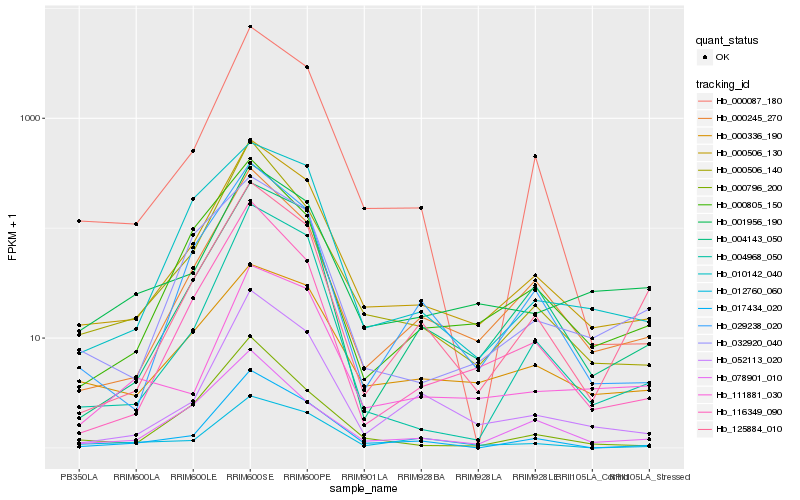
Hb_000506_130 |
0.0 |
- |
- |
DGCR14-related family protein [Populus trichocarpa] |
| 2 |
Hb_000245_270 |
0.0952104476 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 3 |
Hb_000506_140 |
0.1149664996 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |
| 4 |
Hb_052113_020 |
0.1166468856 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 5 |
Hb_017434_020 |
0.1189110994 |
transcription factor |
TF Family: C2H2 |
Zinc finger protein 4 [Populus trichocarpa] |
| 6 |
Hb_029238_020 |
0.12041135 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 7 |
Hb_004143_050 |
0.1204331676 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 8 |
Hb_000087_180 |
0.1266312034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_116349_090 |
0.132490109 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 10 |
Hb_000805_150 |
0.1336421153 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_001956_190 |
0.134188176 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 12 |
Hb_004968_050 |
0.1389896122 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 13 |
Hb_000796_200 |
0.1421284754 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 3 [Jatropha curcas] |
| 14 |
Hb_125884_010 |
0.1441464776 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Jatropha curcas] |
| 15 |
Hb_000336_190 |
0.1445120305 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 16 |
Hb_010142_040 |
0.1477578917 |
- |
- |
PREDICTED: classical arabinogalactan protein 4-like [Jatropha curcas] |
| 17 |
Hb_032920_040 |
0.1481071074 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 18 |
Hb_111881_030 |
0.1517504682 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 19 |
Hb_012760_060 |
0.1529987679 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 20 |
Hb_078901_010 |
0.1543648115 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |