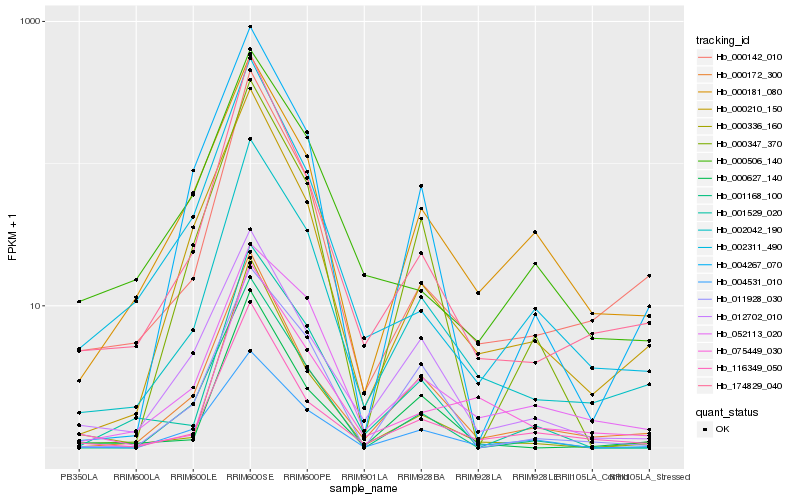
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002042_190 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 2 |
Hb_174829_040 |
0.0925368157 |
- |
- |
PREDICTED: probable calcium-binding protein CML45 [Jatropha curcas] |
| 3 |
Hb_000142_010 |
0.1029468824 |
- |
- |
PREDICTED: uncharacterized protein LOC105632779 [Jatropha curcas] |
| 4 |
Hb_000172_300 |
0.1167400979 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 5 |
Hb_116349_050 |
0.1188297795 |
- |
- |
formin homology 2 domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_052113_020 |
0.1251858418 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 7 |
Hb_000210_150 |
0.1257180972 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 8 |
Hb_004531_010 |
0.1298003142 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 9 |
Hb_000627_140 |
0.130394265 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_075449_030 |
0.1383394275 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 11 |
Hb_001529_020 |
0.1392180072 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_004267_070 |
0.141028945 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 13 |
Hb_000181_080 |
0.142621834 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 14 |
Hb_011928_030 |
0.1456983005 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 15 |
Hb_012702_010 |
0.1457708815 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 16 |
Hb_000347_370 |
0.1465870926 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 17 |
Hb_002311_490 |
0.1469143676 |
- |
- |
PREDICTED: RING-H2 finger protein ATL2 [Jatropha curcas] |
| 18 |
Hb_000336_160 |
0.147963165 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_001168_100 |
0.1524234557 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2-like [Citrus sinensis] |
| 20 |
Hb_000506_140 |
0.1575207047 |
- |
- |
hypothetical protein PRUPE_ppb019425mg [Prunus persica] |