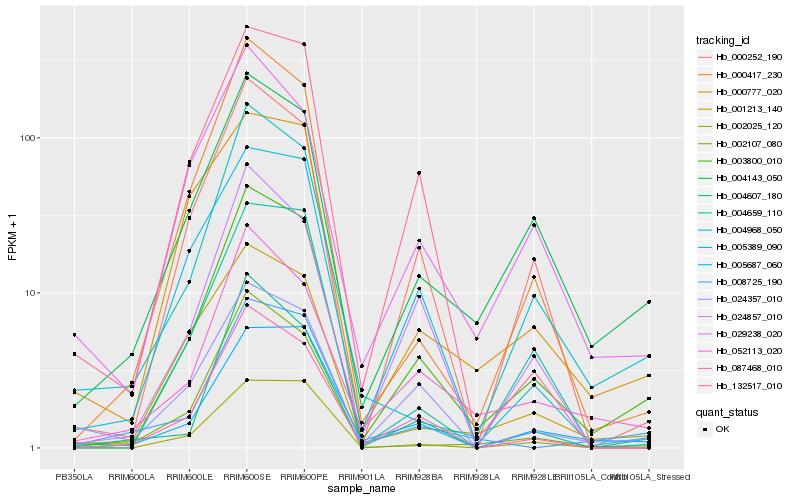
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003800_010 |
0.0 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 2 |
Hb_000252_190 |
0.1148785274 |
- |
- |
PREDICTED: uncharacterized protein LOC105644975 [Jatropha curcas] |
| 3 |
Hb_004143_050 |
0.119829031 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 4 |
Hb_087468_010 |
0.1208822609 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 5 |
Hb_000417_230 |
0.1296532062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_052113_020 |
0.1361376084 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 7 |
Hb_005687_060 |
0.136818323 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004968_050 |
0.1383751166 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 9 |
Hb_001213_140 |
0.141447613 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 10 |
Hb_004607_180 |
0.1436050788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_024357_010 |
0.1442947714 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 12 |
Hb_005389_090 |
0.1448093 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 13 |
Hb_002107_080 |
0.1453320549 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 14 |
Hb_132517_010 |
0.1464707993 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 15 |
Hb_004659_110 |
0.1468926827 |
- |
- |
PREDICTED: BURP domain-containing protein 3-like [Jatropha curcas] |
| 16 |
Hb_024857_010 |
0.1473906287 |
- |
- |
glutaredoxin 2.1 [Hevea brasiliensis] |
| 17 |
Hb_008725_190 |
0.1504811833 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 18 |
Hb_029238_020 |
0.1506339718 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 19 |
Hb_002025_120 |
0.1507847536 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 20 |
Hb_000777_020 |
0.1529024102 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |