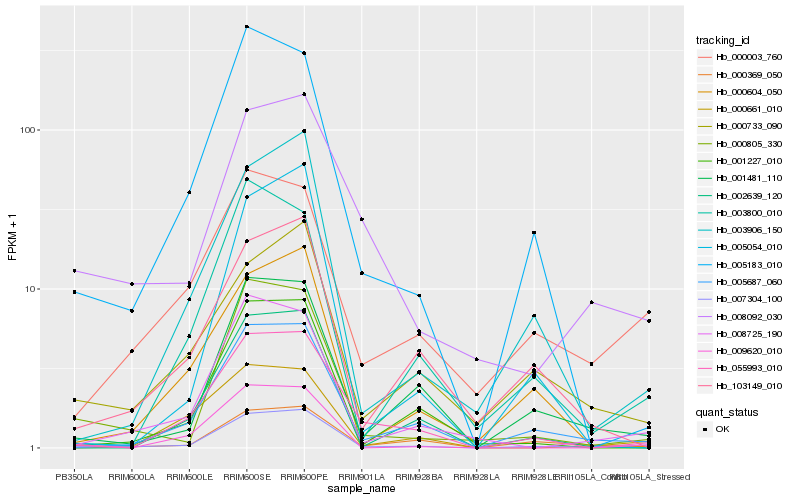
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007304_100 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 2 |
Hb_005687_060 |
0.1233919538 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001481_110 |
0.150844102 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 4 |
Hb_000805_330 |
0.1551621382 |
- |
- |
PREDICTED: ricin-like [Elaeis guineensis] |
| 5 |
Hb_008725_190 |
0.1558027879 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 6 |
Hb_003906_150 |
0.1598320262 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 7 |
Hb_055993_010 |
0.1709817572 |
- |
- |
PREDICTED: uncharacterized protein LOC104883809 [Beta vulgaris subsp. vulgaris] |
| 8 |
Hb_000369_050 |
0.1771167614 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 9 |
Hb_005183_010 |
0.1791818141 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 10 |
Hb_009620_010 |
0.1811852769 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001227_010 |
0.1814118068 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 12 |
Hb_002639_120 |
0.1855885808 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_000003_760 |
0.185847954 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA26-like [Jatropha curcas] |
| 14 |
Hb_008092_030 |
0.1866360241 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 15 |
Hb_000733_090 |
0.1867828887 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 16 |
Hb_000604_050 |
0.1868678343 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 17 |
Hb_005054_010 |
0.1881219579 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_000661_010 |
0.1908734834 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 19 |
Hb_103149_010 |
0.1908937842 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 20 |
Hb_003800_010 |
0.1914184969 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |