| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
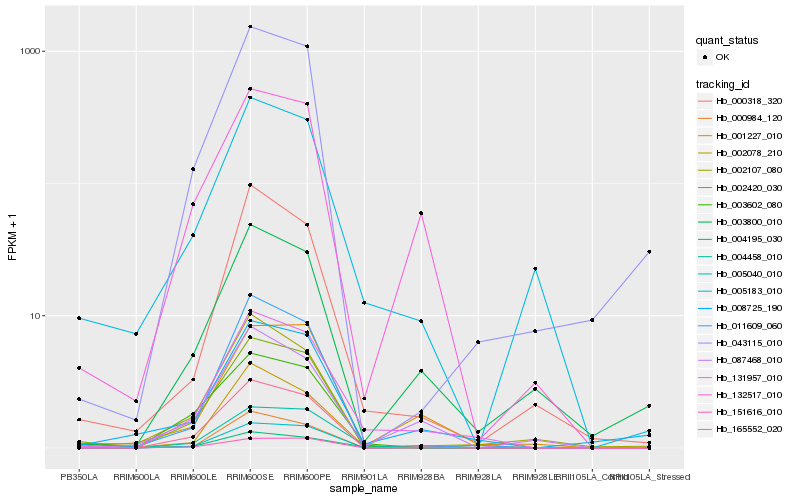
Hb_131957_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 2 |
Hb_004195_030 |
0.1265605229 |
- |
- |
hypothetical protein POPTR_0006s17880g, partial [Populus trichocarpa] |
| 3 |
Hb_004458_010 |
0.1457023744 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 4 |
Hb_000318_320 |
0.1538470463 |
- |
- |
PREDICTED: transmembrane protein 45B [Jatropha curcas] |
| 5 |
Hb_008725_190 |
0.156904006 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 6 |
Hb_132517_010 |
0.1587079947 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 7 |
Hb_001227_010 |
0.1599978979 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 8 |
Hb_002078_210 |
0.1605753442 |
- |
- |
PREDICTED: cytochrome P450 78A5-like [Jatropha curcas] |
| 9 |
Hb_011609_060 |
0.1609360283 |
- |
- |
PREDICTED: cytokinin hydroxylase [Jatropha curcas] |
| 10 |
Hb_000984_120 |
0.162828311 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 11 |
Hb_005183_010 |
0.1643841302 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |
| 12 |
Hb_087468_010 |
0.1660010671 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 13 |
Hb_165552_020 |
0.1666820721 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_003800_010 |
0.1698636315 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 15 |
Hb_003602_080 |
0.1701843655 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 16 |
Hb_151616_010 |
0.1711614552 |
- |
- |
- |
| 17 |
Hb_002420_030 |
0.1722160986 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 18 |
Hb_002107_080 |
0.1735687593 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 19 |
Hb_005040_010 |
0.1741601419 |
- |
- |
hypothetical protein RCOM_1110360 [Ricinus communis] |
| 20 |
Hb_043115_010 |
0.1747300931 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |