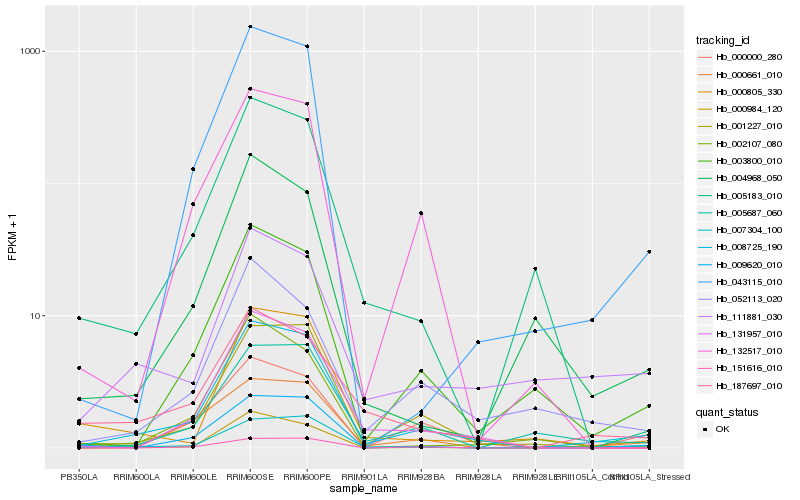
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_190 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 2 |
Hb_003800_010 |
0.1504811833 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 3 |
Hb_001227_010 |
0.1534924684 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 4 |
Hb_007304_100 |
0.1558027879 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 5 |
Hb_131957_010 |
0.156904006 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 6 |
Hb_043115_010 |
0.160538189 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |
| 7 |
Hb_000661_010 |
0.1605547466 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 8 |
Hb_005687_060 |
0.1661378154 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000805_330 |
0.1726195943 |
- |
- |
PREDICTED: ricin-like [Elaeis guineensis] |
| 10 |
Hb_187697_010 |
0.173501503 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 11 |
Hb_132517_010 |
0.1746766708 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like isoform X3 [Populus euphratica] |
| 12 |
Hb_111881_030 |
0.1749862156 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 13 |
Hb_009620_010 |
0.1758825352 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002107_080 |
0.1785748101 |
- |
- |
JHL20J20.3 [Jatropha curcas] |
| 15 |
Hb_000984_120 |
0.1810278167 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_052113_020 |
0.1832444151 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 17 |
Hb_151616_010 |
0.1848130568 |
- |
- |
- |
| 18 |
Hb_000000_280 |
0.1855173301 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004968_050 |
0.1866110055 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 20 |
Hb_005183_010 |
0.1871391882 |
- |
- |
PREDICTED: probable calcium-binding protein CML17 [Malus domestica] |