| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
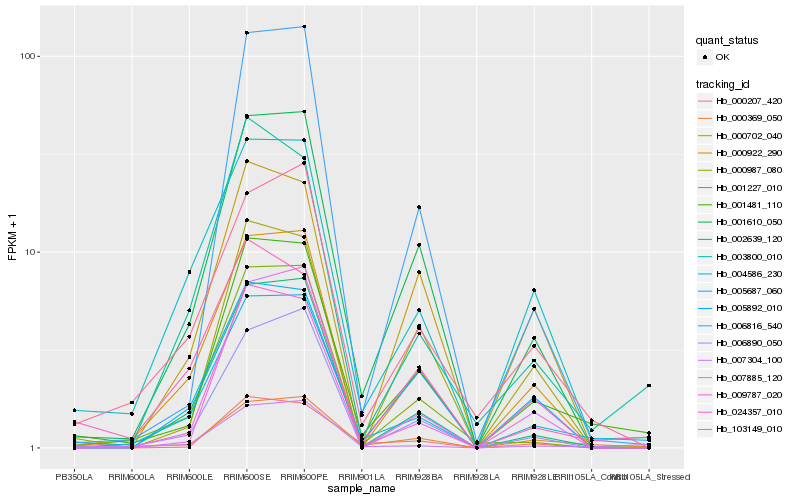
Hb_001481_110 |
0.0 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 2 |
Hb_005687_060 |
0.1000189723 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000369_050 |
0.1061553933 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 4 |
Hb_001610_050 |
0.1286572518 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 5 |
Hb_006816_540 |
0.1434564675 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |
| 6 |
Hb_007304_100 |
0.150844102 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 7 |
Hb_000987_080 |
0.1544648483 |
- |
- |
light-inducible protein atls1, putative [Ricinus communis] |
| 8 |
Hb_000702_040 |
0.156552523 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 9 |
Hb_009787_020 |
0.1580306791 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 10 |
Hb_002639_120 |
0.159913471 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_001227_010 |
0.1611171882 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 12 |
Hb_007885_120 |
0.1629955726 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 13 |
Hb_006890_050 |
0.1634645403 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Jatropha curcas] |
| 14 |
Hb_003800_010 |
0.1645984845 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 15 |
Hb_103149_010 |
0.166131004 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 16 |
Hb_005892_010 |
0.1696299236 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 17 |
Hb_004586_230 |
0.1733908646 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000922_290 |
0.1736352981 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 19 |
Hb_000207_420 |
0.1752519698 |
- |
- |
hypothetical protein POPTR_0019s11600g [Populus trichocarpa] |
| 20 |
Hb_024357_010 |
0.1758706145 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |