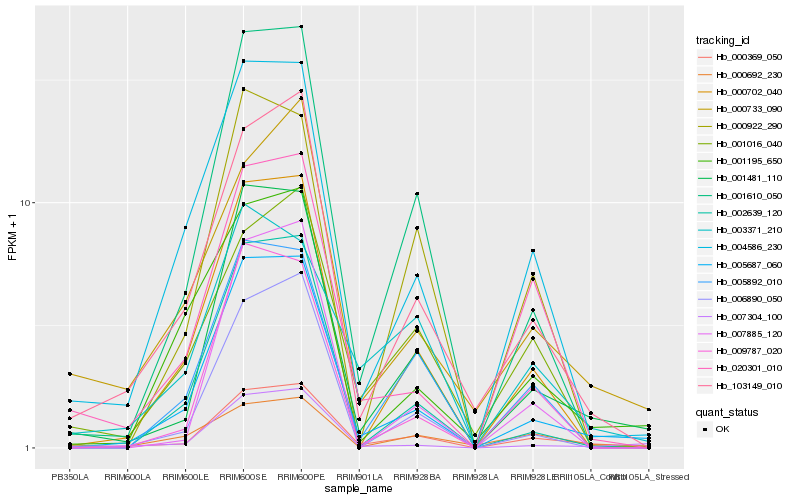
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000369_050 |
0.0 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 2 |
Hb_005687_060 |
0.1047428076 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001481_110 |
0.1061553933 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 4 |
Hb_103149_010 |
0.147473914 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 5 |
Hb_001610_050 |
0.156008815 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 6 |
Hb_000702_040 |
0.1656936448 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 7 |
Hb_005892_010 |
0.1695498881 |
- |
- |
PREDICTED: uncharacterized protein LOC105632168 [Jatropha curcas] |
| 8 |
Hb_004586_230 |
0.1695835813 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007304_100 |
0.1771167614 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 10 |
Hb_009787_020 |
0.179096495 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 11 |
Hb_001016_040 |
0.1821438573 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000733_090 |
0.1828687965 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 13 |
Hb_006890_050 |
0.1835532725 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Jatropha curcas] |
| 14 |
Hb_000692_230 |
0.1836001189 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 15 |
Hb_020301_010 |
0.1865006427 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000922_290 |
0.1884580056 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 17 |
Hb_003371_210 |
0.1898143851 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001195_650 |
0.1905852742 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 19 |
Hb_007885_120 |
0.1920003509 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 20 |
Hb_002639_120 |
0.1920832897 |
- |
- |
unnamed protein product [Vitis vinifera] |