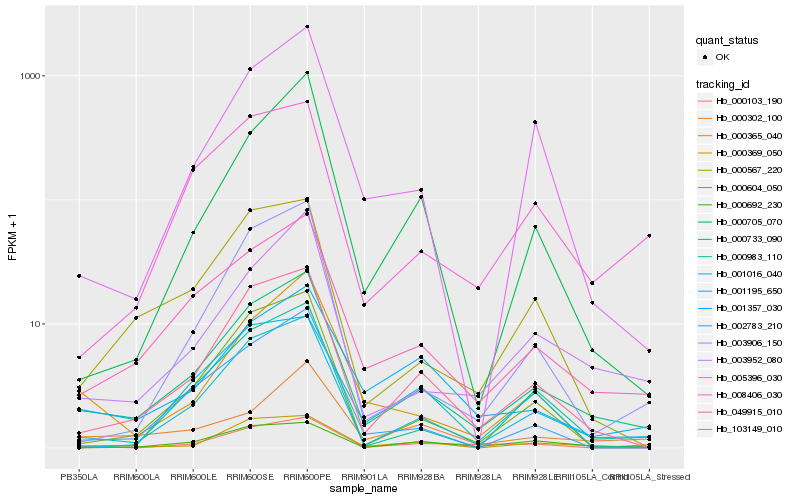
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000733_090 |
0.0 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 2 |
Hb_049915_010 |
0.0934513085 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 3 |
Hb_103149_010 |
0.1163170693 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 4 |
Hb_003952_080 |
0.1331315796 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_000692_230 |
0.154367518 |
- |
- |
phospholipase d delta, putative [Ricinus communis] |
| 6 |
Hb_000604_050 |
0.1575867502 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 7 |
Hb_008406_030 |
0.1629909563 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 8 |
Hb_001016_040 |
0.1642408785 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001357_030 |
0.1655987805 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 10 |
Hb_001195_650 |
0.1656162966 |
- |
- |
PREDICTED: calcium uniporter protein 4, mitochondrial [Jatropha curcas] |
| 11 |
Hb_002783_210 |
0.1656713144 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 12 |
Hb_000567_220 |
0.1667151024 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 13 |
Hb_005396_030 |
0.1669745728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003906_150 |
0.1699891946 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 15 |
Hb_000365_040 |
0.1704982467 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 16 |
Hb_000103_190 |
0.175041146 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 17 |
Hb_000983_110 |
0.1763392517 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 18 |
Hb_000705_070 |
0.1772337027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000302_100 |
0.1792644615 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 20 |
Hb_000369_050 |
0.1828687965 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |