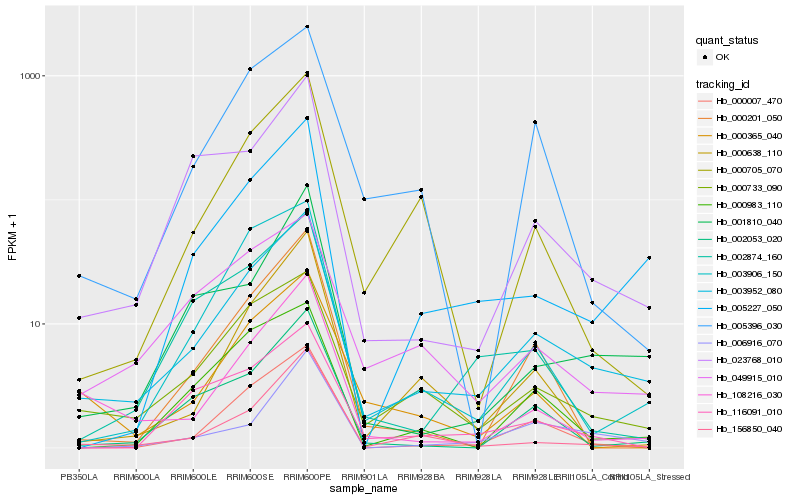
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003952_080 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000733_090 |
0.1331315796 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 3 |
Hb_005227_050 |
0.1577370415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000007_470 |
0.1637338691 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 5 |
Hb_049915_010 |
0.175729123 |
- |
- |
PREDICTED: uncharacterized serine-rich protein C215.13 [Jatropha curcas] |
| 6 |
Hb_108216_030 |
0.176101367 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_023768_010 |
0.1775124771 |
- |
- |
- |
| 8 |
Hb_002053_020 |
0.1778295644 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 9 |
Hb_156850_040 |
0.1790406892 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 10 |
Hb_001810_040 |
0.1819909036 |
- |
- |
- |
| 11 |
Hb_003906_150 |
0.1831136514 |
- |
- |
peptidoglycan-binding LysM domain-containing family protein, partial [Populus trichocarpa] |
| 12 |
Hb_005396_030 |
0.1847487168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000983_110 |
0.1893956773 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 14 |
Hb_000365_040 |
0.1905143757 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 15 |
Hb_002874_160 |
0.1923621284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000201_050 |
0.1942241782 |
- |
- |
- |
| 17 |
Hb_000705_070 |
0.1951253196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_116091_010 |
0.1960041578 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0007s13380g [Populus trichocarpa] |
| 19 |
Hb_000638_110 |
0.1963015747 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 20 |
Hb_006916_070 |
0.1963611219 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1-like [Jatropha curcas] |