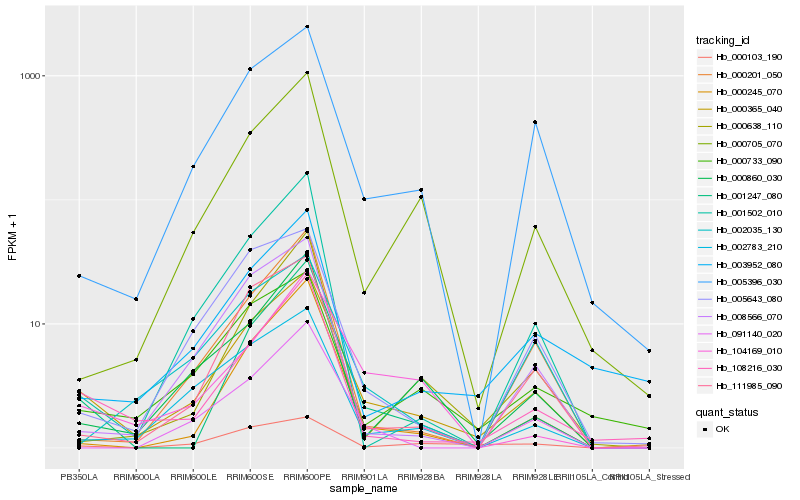
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |
| 2 |
Hb_108216_030 |
0.1482852533 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_005396_030 |
0.1638369685 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000733_090 |
0.1704982467 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 5 |
Hb_001247_080 |
0.1757673397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000860_030 |
0.1758433402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_111985_090 |
0.1774993408 |
- |
- |
hypothetical protein RCOM_1595950 [Ricinus communis] |
| 8 |
Hb_002783_210 |
0.1792209451 |
- |
- |
PREDICTED: uncharacterized protein LOC105635341 [Jatropha curcas] |
| 9 |
Hb_000705_070 |
0.1877766919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008566_070 |
0.1890790601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000245_070 |
0.1891734281 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 12 |
Hb_005643_080 |
0.1893085878 |
- |
- |
hypothetical protein POPTR_0001s12740g [Populus trichocarpa] |
| 13 |
Hb_003952_080 |
0.1905143757 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000103_190 |
0.1924011456 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 15 |
Hb_000201_050 |
0.1938553033 |
- |
- |
- |
| 16 |
Hb_104169_010 |
0.1952441256 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 17 |
Hb_001502_010 |
0.1962374093 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 18 |
Hb_000638_110 |
0.201293801 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 19 |
Hb_002035_130 |
0.2026079249 |
- |
- |
plasma membrane H+-ATPase [Hevea brasiliensis] |
| 20 |
Hb_091140_020 |
0.202814247 |
- |
- |
PREDICTED: uncharacterized protein LOC105640903 [Jatropha curcas] |