| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
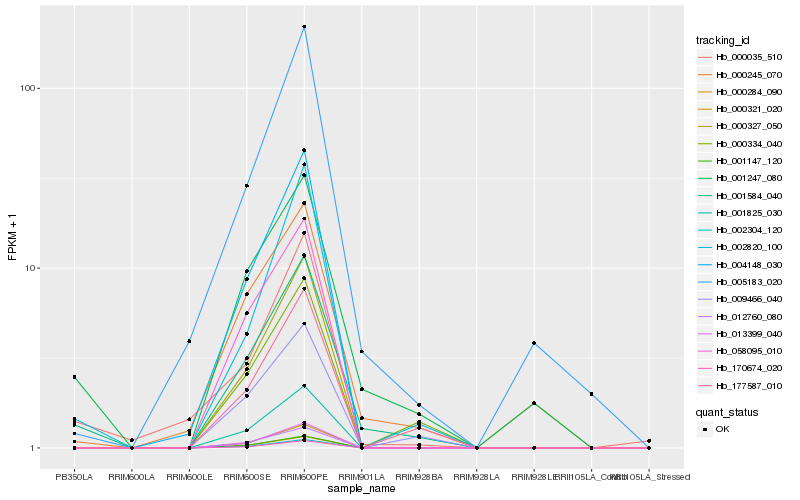
Hb_001584_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105125166 [Populus euphratica] |
| 2 |
Hb_001247_080 |
0.1093424669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002820_100 |
0.1547282911 |
- |
- |
hypothetical protein OsI_33031 [Oryza sativa Indica Group] |
| 4 |
Hb_000245_070 |
0.1662890501 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 5 |
Hb_009466_040 |
0.1671589226 |
- |
- |
hypothetical protein POPTR_0002s10890g [Populus trichocarpa] |
| 6 |
Hb_001147_120 |
0.1691849362 |
- |
- |
hypothetical protein JCGZ_14802 [Jatropha curcas] |
| 7 |
Hb_001825_030 |
0.1692776205 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 8 |
Hb_000284_090 |
0.1693010766 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 9 |
Hb_000321_020 |
0.1701225518 |
- |
- |
PREDICTED: uncharacterized protein LOC104891638 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_177587_010 |
0.170573535 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 11 |
Hb_002304_120 |
0.1708653095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000035_510 |
0.1711321417 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 13 |
Hb_012760_080 |
0.1716808322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000327_050 |
0.172031565 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.7-like isoform X1 [Populus euphratica] |
| 15 |
Hb_000334_040 |
0.1723146412 |
- |
- |
hypothetical protein POPTR_0001s41860g [Populus trichocarpa] |
| 16 |
Hb_013399_040 |
0.1733181068 |
- |
- |
hypothetical protein JCGZ_24713 [Jatropha curcas] |
| 17 |
Hb_058095_010 |
0.1742301534 |
- |
- |
PREDICTED: uncharacterized protein LOC105109431 [Populus euphratica] |
| 18 |
Hb_170674_020 |
0.175396349 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 19 |
Hb_004148_030 |
0.1756826037 |
- |
- |
PREDICTED: protein RAFTIN 1B-like [Gossypium raimondii] |
| 20 |
Hb_005183_020 |
0.175821593 |
- |
- |
hypothetical protein JCGZ_04285 [Jatropha curcas] |