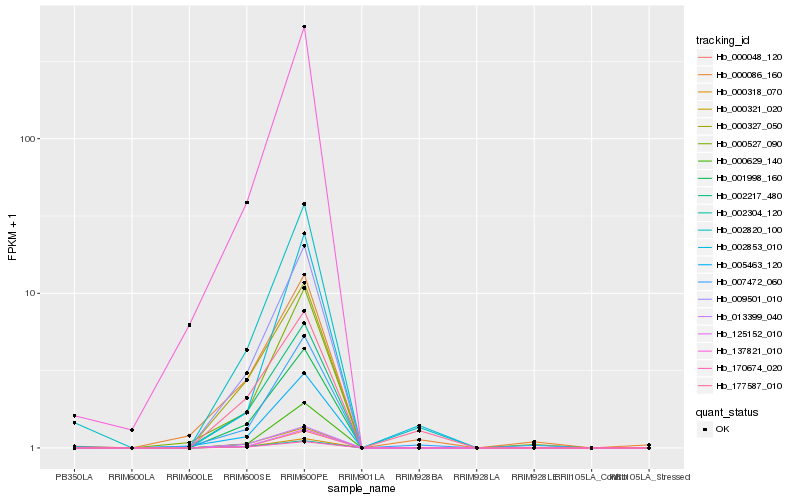
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002820_100 |
0.0 |
- |
- |
hypothetical protein OsI_33031 [Oryza sativa Indica Group] |
| 2 |
Hb_007472_060 |
0.0983381757 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_125152_010 |
0.1016436848 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 4 |
Hb_009501_010 |
0.1018369753 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 5 |
Hb_000048_120 |
0.1018935082 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 6 |
Hb_005463_120 |
0.1033657874 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 7 |
Hb_001998_160 |
0.10757745 |
- |
- |
PREDICTED: origin recognition complex subunit 1-like [Populus euphratica] |
| 8 |
Hb_000629_140 |
0.1090575495 |
- |
- |
hypothetical protein SORBIDRAFT_04g013083 [Sorghum bicolor] |
| 9 |
Hb_000318_070 |
0.1145884292 |
- |
- |
PREDICTED: peroxidase 10-like [Jatropha curcas] |
| 10 |
Hb_002217_480 |
0.1151893513 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 11 |
Hb_137821_010 |
0.1155619809 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 12 |
Hb_170674_020 |
0.1174914002 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 13 |
Hb_013399_040 |
0.122267121 |
- |
- |
hypothetical protein JCGZ_24713 [Jatropha curcas] |
| 14 |
Hb_000327_050 |
0.1260993037 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.7-like isoform X1 [Populus euphratica] |
| 15 |
Hb_002853_010 |
0.1286902955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000527_090 |
0.1303610943 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 17 |
Hb_002304_120 |
0.1306437011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000086_160 |
0.1345094188 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 19 |
Hb_000321_020 |
0.1346084302 |
- |
- |
PREDICTED: uncharacterized protein LOC104891638 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_177587_010 |
0.1360694678 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |