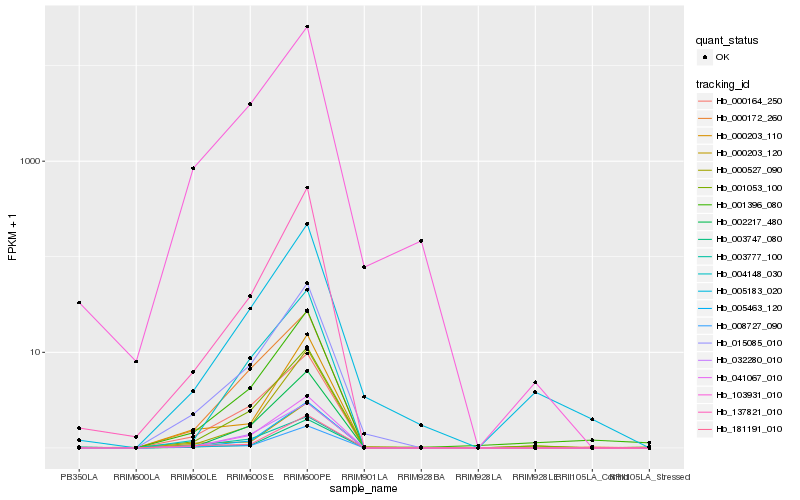
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015085_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 2 |
Hb_041067_010 |
0.0687176304 |
- |
- |
hypothetical protein JCGZ_00780 [Jatropha curcas] |
| 3 |
Hb_103931_010 |
0.0713155408 |
- |
- |
- |
| 4 |
Hb_001053_100 |
0.0819571792 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 5 |
Hb_032280_010 |
0.0921622534 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000164_250 |
0.0941447064 |
- |
- |
hypothetical protein CISIN_1g040645mg [Citrus sinensis] |
| 7 |
Hb_003747_080 |
0.0964437956 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 8 |
Hb_181191_010 |
0.0972985698 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_000172_260 |
0.0991348041 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 10 |
Hb_005463_120 |
0.0993610383 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 11 |
Hb_002217_480 |
0.099486922 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 12 |
Hb_137821_010 |
0.1008603908 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 13 |
Hb_005183_020 |
0.1023229225 |
- |
- |
hypothetical protein JCGZ_04285 [Jatropha curcas] |
| 14 |
Hb_004148_030 |
0.1029521831 |
- |
- |
PREDICTED: protein RAFTIN 1B-like [Gossypium raimondii] |
| 15 |
Hb_008727_090 |
0.1057192114 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 16 |
Hb_003777_100 |
0.1095481974 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 17 |
Hb_000203_120 |
0.1102206724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000527_090 |
0.1112288553 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_000203_110 |
0.1127753026 |
- |
- |
- |
| 20 |
Hb_001396_080 |
0.1142469515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |