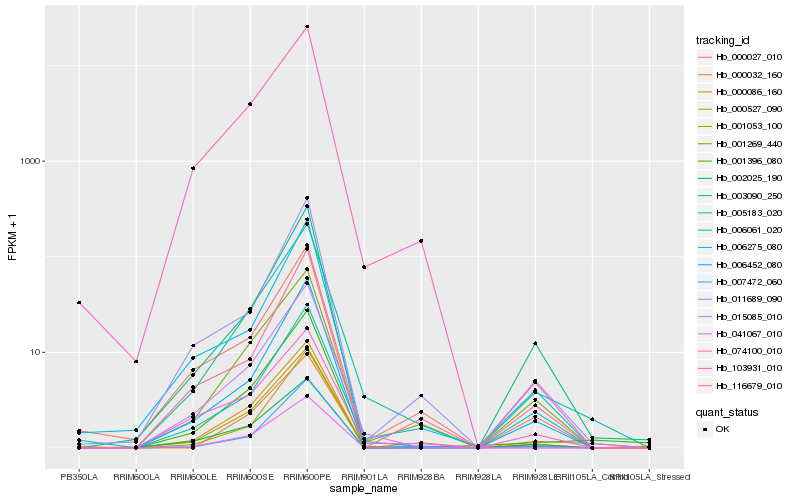
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005183_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_04285 [Jatropha curcas] |
| 2 |
Hb_001053_100 |
0.0837615746 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 3 |
Hb_001269_440 |
0.0932764411 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 4 |
Hb_001396_080 |
0.1004250673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_015085_010 |
0.1023229225 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 6 |
Hb_000086_160 |
0.1023998513 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 7 |
Hb_000032_160 |
0.1026911283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006061_020 |
0.1084520512 |
transcription factor |
TF Family: bZIP |
bZIP family protein [Populus trichocarpa] |
| 9 |
Hb_103931_010 |
0.1097369952 |
- |
- |
- |
| 10 |
Hb_007472_060 |
0.115136418 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_006452_080 |
0.1153719425 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 12 |
Hb_000027_010 |
0.1158281299 |
- |
- |
- |
| 13 |
Hb_002025_190 |
0.118413161 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 14 |
Hb_003090_250 |
0.1189462464 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_011689_090 |
0.1189687076 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 16 |
Hb_006275_080 |
0.1197043784 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 17 |
Hb_041067_010 |
0.1197864208 |
- |
- |
hypothetical protein JCGZ_00780 [Jatropha curcas] |
| 18 |
Hb_000527_090 |
0.1198989059 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 19 |
Hb_074100_010 |
0.1215048158 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 20 |
Hb_116679_010 |
0.1274178861 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |