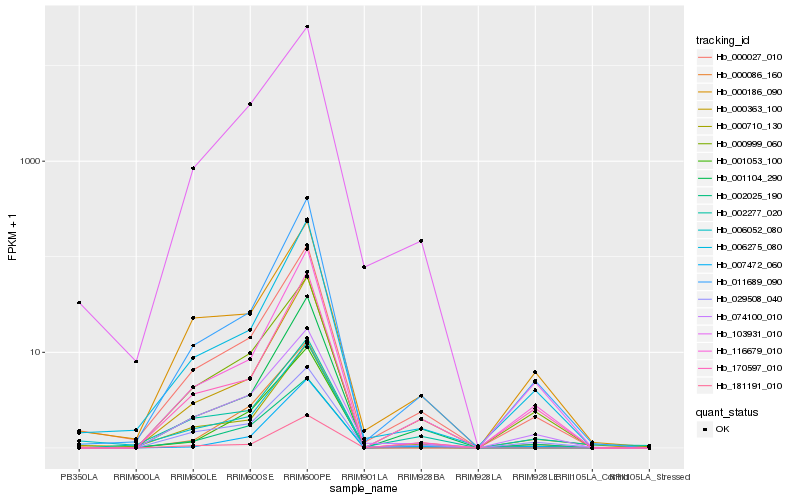
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_006275_080 |
0.0687987112 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 3 |
Hb_011689_090 |
0.0768943087 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 4 |
Hb_000710_130 |
0.0785154379 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 5 |
Hb_116679_010 |
0.0837240983 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 6 |
Hb_000186_090 |
0.0892144327 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 7 |
Hb_006052_080 |
0.0905269844 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH140-like [Populus euphratica] |
| 8 |
Hb_103931_010 |
0.0910605423 |
- |
- |
- |
| 9 |
Hb_002025_190 |
0.0941427404 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 10 |
Hb_000086_160 |
0.0943886979 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 11 |
Hb_000363_100 |
0.0944958341 |
- |
- |
- |
| 12 |
Hb_001104_290 |
0.0960840975 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 13 |
Hb_029508_040 |
0.0991956306 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 14 |
Hb_170597_010 |
0.0993204635 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 15 |
Hb_074100_010 |
0.1001500158 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 16 |
Hb_001053_100 |
0.1009033993 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 17 |
Hb_000999_060 |
0.1076942215 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |
| 18 |
Hb_007472_060 |
0.1103931039 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_181191_010 |
0.1112697148 |
- |
- |
kinase, putative [Ricinus communis] |
| 20 |
Hb_002277_020 |
0.1120852418 |
- |
- |
Uncharacterized protein TCM_002409 [Theobroma cacao] |