| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
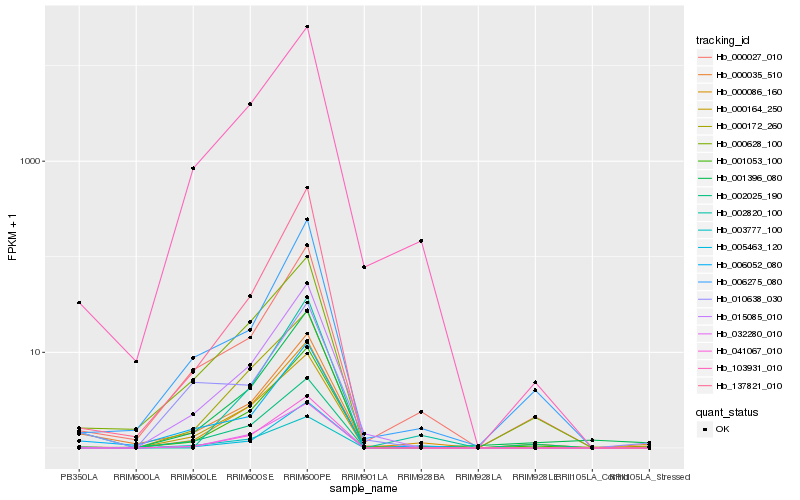
Hb_000035_510 |
0.0 |
- |
- |
PREDICTED: protein GLUTAMINE DUMPER 5-like [Jatropha curcas] |
| 2 |
Hb_006052_080 |
0.0899619179 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH140-like [Populus euphratica] |
| 3 |
Hb_001053_100 |
0.1211029308 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 4 |
Hb_103931_010 |
0.1218873226 |
- |
- |
- |
| 5 |
Hb_005463_120 |
0.1222326927 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 6 |
Hb_000027_010 |
0.1305013999 |
- |
- |
- |
| 7 |
Hb_000628_100 |
0.1337742315 |
- |
- |
hypothetical protein MTR_1g019660 [Medicago truncatula] |
| 8 |
Hb_000086_160 |
0.1358277418 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 9 |
Hb_015085_010 |
0.1387991062 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 10 |
Hb_041067_010 |
0.1471705957 |
- |
- |
hypothetical protein JCGZ_00780 [Jatropha curcas] |
| 11 |
Hb_001396_080 |
0.1493789716 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010638_030 |
0.1517968208 |
- |
- |
oleosin1 [Plukenetia volubilis] |
| 13 |
Hb_000164_250 |
0.1530069334 |
- |
- |
hypothetical protein CISIN_1g040645mg [Citrus sinensis] |
| 14 |
Hb_006275_080 |
0.1539912851 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 15 |
Hb_032280_010 |
0.1554210875 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 16 |
Hb_137821_010 |
0.1559750479 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 17 |
Hb_000172_260 |
0.1563958087 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 18 |
Hb_002820_100 |
0.1577199564 |
- |
- |
hypothetical protein OsI_33031 [Oryza sativa Indica Group] |
| 19 |
Hb_002025_190 |
0.159903336 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 20 |
Hb_003777_100 |
0.1600284831 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |