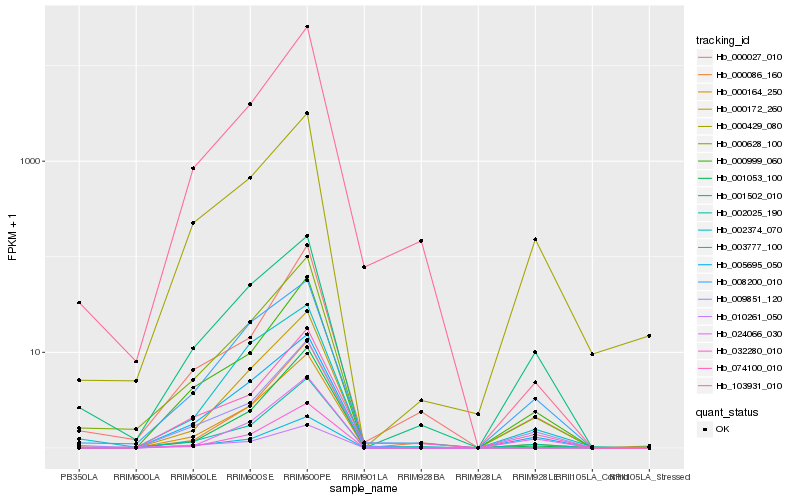
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000628_100 |
0.0 |
- |
- |
hypothetical protein MTR_1g019660 [Medicago truncatula] |
| 2 |
Hb_002025_190 |
0.0779118914 |
- |
- |
PREDICTED: transcription factor bHLH111 [Jatropha curcas] |
| 3 |
Hb_000999_060 |
0.0944159313 |
- |
- |
hypothetical protein POPTR_0018s11350g [Populus trichocarpa] |
| 4 |
Hb_000164_250 |
0.0958261536 |
- |
- |
hypothetical protein CISIN_1g040645mg [Citrus sinensis] |
| 5 |
Hb_003777_100 |
0.0971246411 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 6 |
Hb_005695_050 |
0.0980621054 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 7 |
Hb_009851_120 |
0.1005900267 |
- |
- |
PREDICTED: RING-H2 zinc finger protein RHA4a [Jatropha curcas] |
| 8 |
Hb_001053_100 |
0.1058838034 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 9 |
Hb_000172_260 |
0.1081696144 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 10 |
Hb_032280_010 |
0.1084626616 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 11 |
Hb_103931_010 |
0.1090734671 |
- |
- |
- |
| 12 |
Hb_000429_080 |
0.1158875855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000086_160 |
0.1196368403 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 14 |
Hb_000027_010 |
0.1198029677 |
- |
- |
- |
| 15 |
Hb_074100_010 |
0.1199238383 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 16 |
Hb_010261_050 |
0.1203542351 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_024066_030 |
0.1214508789 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 18 |
Hb_002374_070 |
0.1219562429 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_001502_010 |
0.123926524 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 20 |
Hb_008200_010 |
0.1248977252 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |