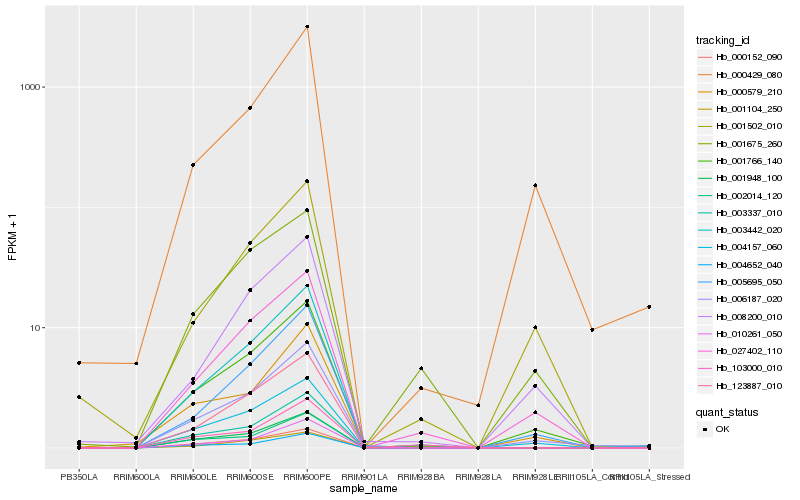
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001766_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 2 |
Hb_006187_020 |
0.0512633281 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Populus euphratica] |
| 3 |
Hb_027402_110 |
0.0779998359 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 4 |
Hb_005695_050 |
0.0951108428 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 5 |
Hb_004157_060 |
0.098452302 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase protein 22-like [Jatropha curcas] |
| 6 |
Hb_000429_080 |
0.1071817259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008200_010 |
0.1085576412 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003442_020 |
0.1118041803 |
- |
- |
ccd1, putative [Ricinus communis] |
| 9 |
Hb_003337_010 |
0.1152965334 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001502_010 |
0.1170379467 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 11 |
Hb_000579_210 |
0.1174348919 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 12 |
Hb_001948_100 |
0.1180559008 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000152_090 |
0.1186235372 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_123887_010 |
0.1224281493 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 15 |
Hb_001675_260 |
0.1224725276 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 16 |
Hb_103000_010 |
0.1227346222 |
- |
- |
hypothetical protein POPTR_0007s02210g [Populus trichocarpa] |
| 17 |
Hb_010261_050 |
0.1235157458 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_001104_250 |
0.1246845602 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Ricinus communis] |
| 19 |
Hb_004652_040 |
0.125334745 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_002014_120 |
0.1275639633 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP8 [Populus euphratica] |