| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
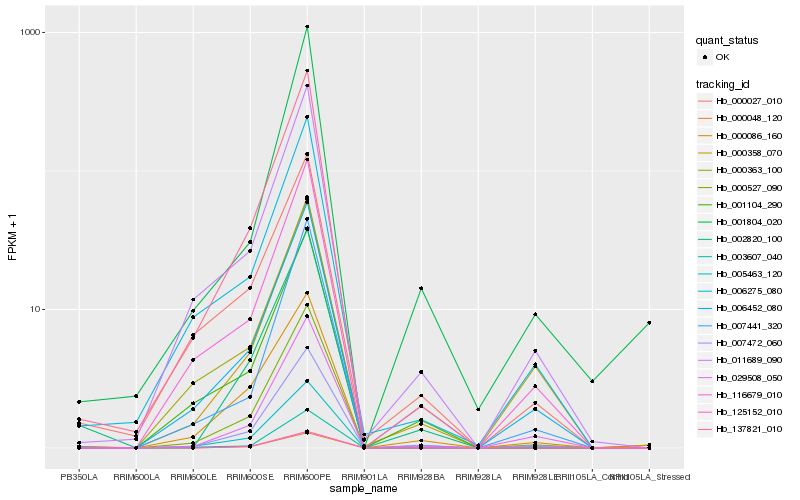
Hb_007472_060 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000527_090 |
0.0797293573 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_011689_090 |
0.080838713 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 4 |
Hb_029508_050 |
0.0810426984 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like isoform X2 [Musa acuminata subsp. malaccensis] |
| 5 |
Hb_116679_010 |
0.0840743506 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 6 |
Hb_006452_080 |
0.0888765882 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 7 |
Hb_003607_040 |
0.0923452501 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_002820_100 |
0.0983381757 |
- |
- |
hypothetical protein OsI_33031 [Oryza sativa Indica Group] |
| 9 |
Hb_000086_160 |
0.0987897929 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 10 |
Hb_137821_010 |
0.0988140923 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 11 |
Hb_006275_080 |
0.1004186654 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 12 |
Hb_000358_070 |
0.1027098464 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 13 |
Hb_007441_320 |
0.1042601258 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 14 |
Hb_001104_290 |
0.1062081158 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 15 |
Hb_005463_120 |
0.1084939843 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 16 |
Hb_000363_100 |
0.109532742 |
- |
- |
- |
| 17 |
Hb_000027_010 |
0.1103931039 |
- |
- |
- |
| 18 |
Hb_001804_020 |
0.1120420765 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 19 |
Hb_000048_120 |
0.112321904 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 20 |
Hb_125152_010 |
0.1123497513 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |