| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
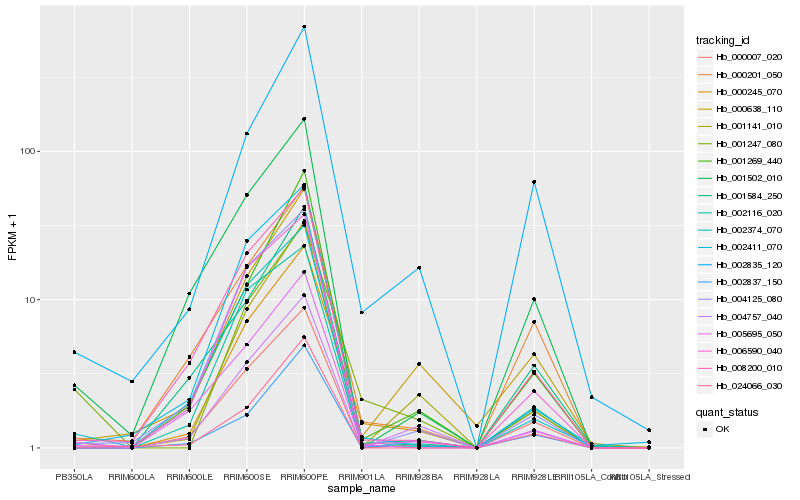
Hb_000245_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105141276 [Populus euphratica] |
| 2 |
Hb_004757_040 |
0.1122946404 |
- |
- |
- |
| 3 |
Hb_000007_020 |
0.1149292909 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 4 |
Hb_001269_440 |
0.1180878927 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |
| 5 |
Hb_002835_120 |
0.1196749242 |
- |
- |
PREDICTED: uncharacterized protein LOC105640304 [Jatropha curcas] |
| 6 |
Hb_006590_040 |
0.1215546251 |
- |
- |
amino acid permease, putative [Ricinus communis] |
| 7 |
Hb_008200_010 |
0.1242792859 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004125_080 |
0.1272202924 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Populus euphratica] |
| 9 |
Hb_001247_080 |
0.1292471432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000638_110 |
0.129457156 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0007s04250g [Populus trichocarpa] |
| 11 |
Hb_001141_010 |
0.1312688676 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 12 |
Hb_002374_070 |
0.1332583597 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_002411_070 |
0.1363967985 |
- |
- |
PREDICTED: WAT1-related protein At5g07050 [Jatropha curcas] |
| 14 |
Hb_002116_020 |
0.1386827788 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 35 [Jatropha curcas] |
| 15 |
Hb_005695_050 |
0.1391929348 |
- |
- |
PREDICTED: sulfate transporter 1.3-like [Jatropha curcas] |
| 16 |
Hb_000201_050 |
0.1395303158 |
- |
- |
- |
| 17 |
Hb_001584_250 |
0.1401425551 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 18 |
Hb_024066_030 |
0.1403568808 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 19 |
Hb_002837_150 |
0.1403979893 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 20 |
Hb_001502_010 |
0.1430311908 |
- |
- |
hypothetical protein JCGZ_04288 [Jatropha curcas] |